-
Notifications
You must be signed in to change notification settings - Fork 1
Deidentification Process
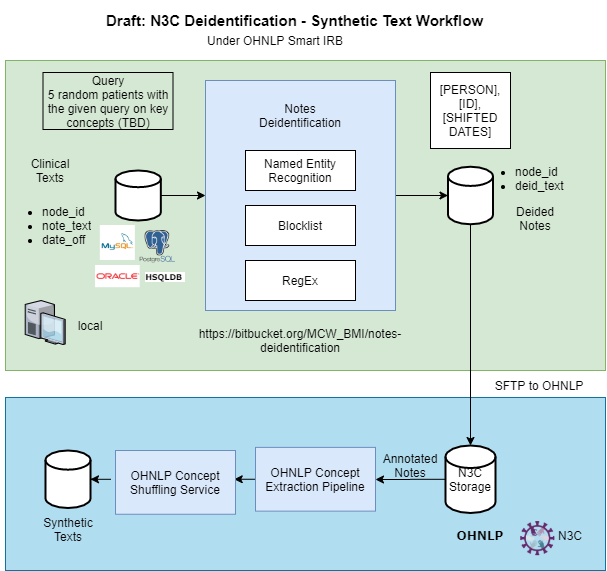
We would like to document the steps for the deidentification of clinical narratives for N3C.
- 5 inpatients and 5 outpatients with research authorizations
- 2 weeks before AND 4 weeks after the lab order date of the first positive COVID-19 results
- Select only the notes with more than 1000 characters
- Up to 20 notes from each patient
- If a patient has more than 20 notes, please stratify the collection based on note date (do NOT cut them off by the date)
- Java 8
- A database connection with login credentials
- Currently with supports of
HSQL,MySQL,PostgresQLandOracle DB.
- Currently with supports of
- The corresponding JDBC Driver
We are using the notes-deidentification tool developed by the Medical College of Wisconsin Biomedical Informatics team.
Obtain the library of notes-deidentification via:
git clone https://bitbucket.org/MCW_BMI/notes-deidentification-standalone.git
Regarding the input from the database, the minimal requirements of the columns for the deid algorithm includes id, note_text and date_off defined as:
-
id: unique note identifier -
note_text: text to be de-identified. -
date_off: numeric offset for date de-identification, e.g., -15 t0 +15. Set to zero to not de-id dates.
Given the name of the output table, the notes with PHI removed will be generated as the deid_note_text column.
Run runDid.sh with the filled variables and the output notes can be found at the table named by deidnotestablename.
If any of the PHI mentions that were not detected or removed by the notes-deidentification need to be manually removed. Depending on the content of PHI, the mentions should be replaced with the following tags:
[MISS-PHI-NAME][MISS-PHI-DATE][MISS-PHI-ADDRESS][MISS-PHI-ID][MISS-PHI-OTHERS]
This site including its contents of concept glossary, risk factors and architecture is a demonstration of work-in-progress of the N3C and OHNLP groups. The contents of the page is under Apache License Version 2.0.