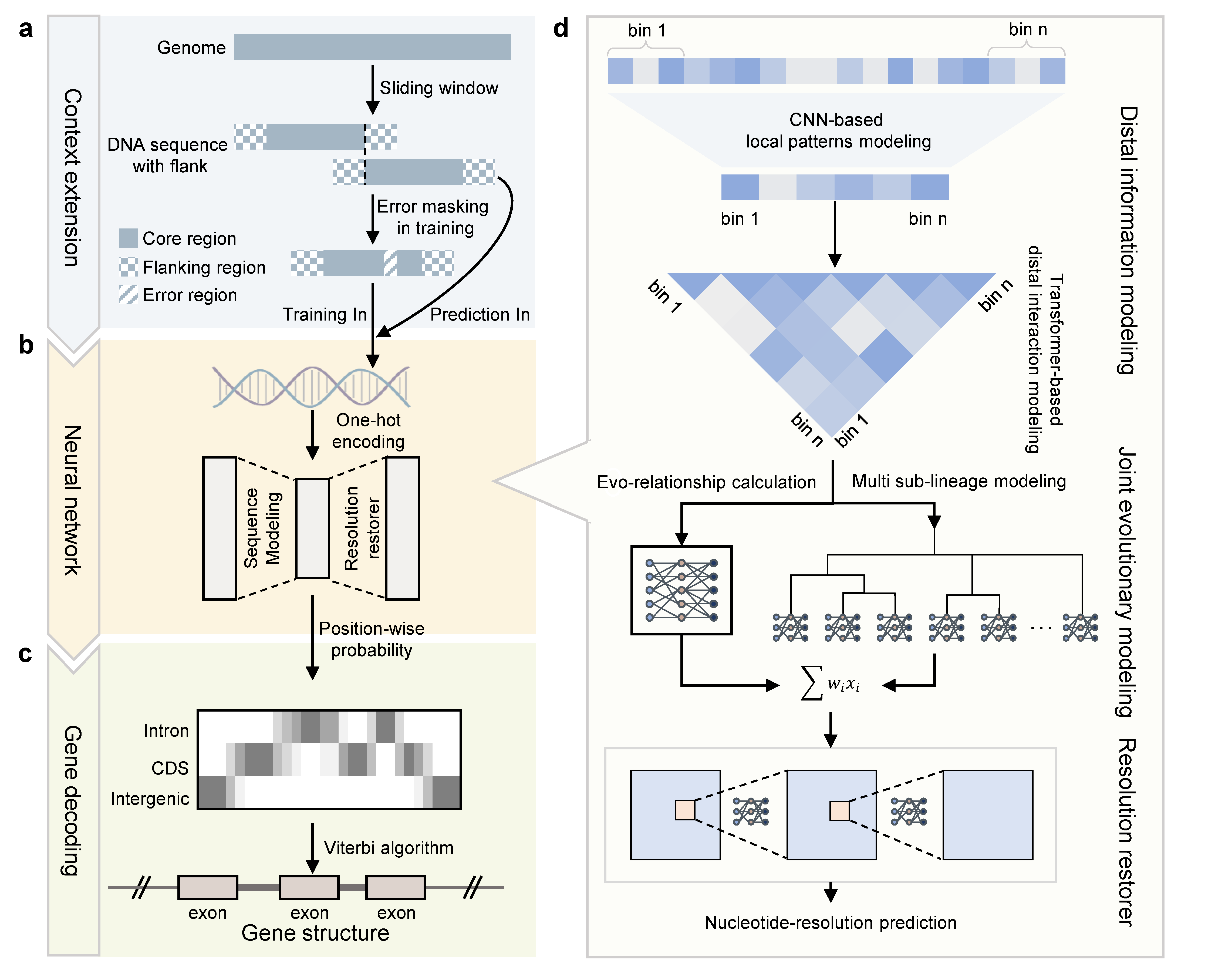
ANNEVO is a deep learning-based ab initio gene annotation method for understanding genome function. ANNEVO is capable of modeling distal sequence information and joint evolutionary relationships across diverse species directly from genomes.
While ANNEVO is released under the GPL-3.0 license, its use is restricted to non-commercial purposes only. This includes use by academic institutions, government agencies, and non-profit or not-for-profit organizations.
A commercial license of the software is available and licensed through Xi’an Jiaotong University. For commercial use or licensing inquiries, please contact: Pengyu Zhang (pengyuzhang@stu.xjtu.edu.cn) or Kai Ye (kaiye@xjtu.edu.cn).
We recommend using the conda virtual environment to install ANNEVO (Platform: Linux).
# Get the source code
git clone https://github.com/xjtu-omics/ANNEVO.git
cd ANNEVO
# Create a conda environment for ANNEVO
conda create -n ANNEVO python=3.6
# Activate conda environment
conda activate ANNEVO
# To use GPU acceleration properly, we recommend installing PyTorch using the official installation
# commands provided by PyTorch (https://pytorch.org/get-started/previous-versions/).
# Select the appropriate command based on your CUDA version to install PyTorch version 1.10.
# Or directly use `pytorch-cuda` to automatically install the appropriate `cudatoolkit`.
# For example, if the CUDA version is not lower than 11.8, you can use the following command:
conda install pytorch=1.10 torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia
# Install other packages
pip install -r requirements.txtCheck if CUDA is available:
import torch
print(torch.cuda.is_available())python annotation.py --genome path_to_genome --model_path path_to_model --output path_to_gff --threads 48We strongly recommend utilizing more CPU cores by adjusting threads when sufficient computational resources are available, as this will significantly accelerate the computation. If your GPU environment has limited CPU resources, you can also use the step-by-step execution mode.
Note: ANNEVO automatically supports use in a multi-GPU environment. If GPU resources are insufficient, you can adjust it by --batch_size. For example, adding the parameter --batch_size 8 only requires about 3G GPU memory.
Typically, deep learning is conducted in environments equipped with GPU resources, where CPU resources are often limited. However, decoding gene structures usually requires substantial CPU resources. To address this, we provide a segmented execution approach, allowing users to flexibly switch between computational nodes/environments with different resources.
Stage 1: Predicting three types of information for each nucleotide (recommended to be performed on environments with abundant GPU resources).
Stage 2: Decoding the three types of information into biologically valid gene structures (recommended to be performed on environments with abundant CPU resources).
# Nucleotide prediction
python prediction.py --genome path_to_genome --model_path path_to_model --model_prediction_path path_to_save_predction
# Gene structure decoding
python decoding.py --genome path_to_genome --model_prediction_path path_to_save_predction --output path_to_gff --threads 48 The demo data located at './example'.
Arabidopsis_chr4_genome.fna: Genome sequence of chromosome 4 of Arabidopsis thaliana.
Arabidopsis_chr4_annotation.gff: RefSeq annotation of chromosome 4 of Arabidopsis thaliana.
# One-step Execution
python annotation.py --genome example/Arabidopsis_chr4_genome.fna --model_path ANNEVO_model/ANNEVO_Embryophyta.pt --output gff_result/Arabidopsis_chr4_annotation.gff --threads 48
# Step-by-step Execution
python prediction.py --genome example/Arabidopsis_chr4_genome.fna --model_path ANNEVO_model/ANNEVO_Embryophyta.pt --model_prediction_path prediction_result/Arabidopsis_chr4/
python decoding.py --genome example/Arabidopsis_chr4_genome.fna --model_prediction_path prediction_result/Arabidopsis_chr4 --output gff_result/Arabidopsis_chr4_annotation.gff --threads 48When you need to incorporate additional species or retrain ANNEVO on a specific clade, you can follow the steps below:
# Filter out duplicated gene IDs and other issues that may cause parsing errors in the Biopython package
python src/filter_wrong_record.py --input_file path_to_annotation --output_file path_to_filtered_annotation
# Convert the genome sequence and annotation into H5 data for model training.
python generate_datasets.py --genome path_to_genome --annotation path_to_filtered_annotation --output_file path_to_h5_data
# Train deep learning model
python model_train.py --train_list path_to_train_species_list --val_list path_to_val_species_list --model_save_path path_to_saved_model --h5_path path_to_h5_dataThe path_to_h5_data directory should contain all H5-formatted data files used for both the training and validation sets.
The train_species_list and val_species_list files are plain text files that specify which species to use by matching the corresponding H5 filenames.
Model parameter files are saved with the .pt file extension.
For example, if you use ten species for training and name the corresponding H5 files as species_name_1 through species_name_10, then train_species_list should be a .txt file containing the following lines:
species_name_1
species_name_2
...
species_name_10
In cases where closely related species are limited or unavailable for the target genome, one of ANNEVO’s five main trained models can be selected as a starting point for fine-tuning.
# Filter out duplicated gene IDs and other issues that may cause parsing errors in the Biopython package
python src/filter_wrong_record.py --input_file path_to_annotation --output_file path_to_filtered_annotation
# Convert the genome sequence and annotation into H5 data for model training.
python generate_datasets.py --genome path_to_genome --annotation path_to_filtered_annotation --output_file path_to_h5_data
# Fine tuning deep learning model
python fine_tune.py --fine_tune_species_list path_to_species_list --model_path path_to_model --model_save_path path_to_fine_tuned_model --h5_path path_to_h5_dataThe path_to_h5_data, species_list and model parameter files are the same as those described in the Re-train ANNEVO section.
If you have any questions, please feel free to contact: pengyuzhang@stu.xjtu.edu.cn