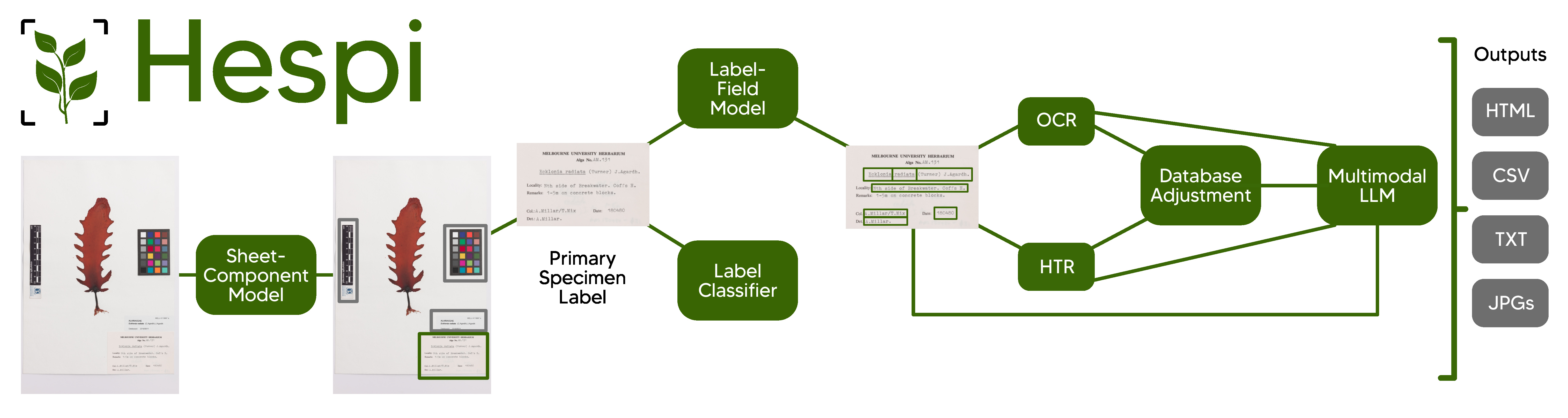
HErbarium Specimen sheet PIpeline
Hespi takes images of specimen sheets from herbaria and first detects the various components of the sheet.
To read more about Hespi, see our paper in BioScience or the article in The Conversation.

Hespi first takes a specimen sheet and detects the various components of it using the Sheet-Component Model. Then any full database label detected is cropped and this is given to the Label-Field Model which detects different textual fields written on the label. A Label Classifier is also used to determine the type of text written on the label. If it is printed or typewritten, then the text of each field is given to an Optical Character Recognition (OCR) engine and if there is handwriting, then each field is given to the Handwritten Text Recognition (HTR) engine. The recognized text is then corrected using a multimodal Large Language Model (LLM). Finally, the result of the fields is post-processed before being written into an HTML report, a CSV file and text files.
The stages of the pipeline are explained in the documentation for the pipeline and our poster.
Install hespi using pip:
pip install hespiThe first time it runs, it will download the required model weights from the internet.
It is recommended that you also install Tesseract so that this can be used in the text recognition part of the pipeline.
To install the development version, see the documentation for contributing.
To run the pipeline, use the executable hespi and give it any number of images:
hespi image1.jpg image2.jpgBy default the output will go to a directory called hespi-output.
You can set the output directory with the command with the --output-dir argument:
hespi images/*.tif --output-dir ./hespi-outputThe detected components and text fields will be cropped and stored in the output directory.
There will also be a CSV file with the filename hespi-results.csv in the output directory with the text recognition results for any primary specimen labels found.
By default hespi will use OpenAI's gpt-4o large language model (LLM) in the pipeline to produce the final results.
If you wish to use a different model from OpenAI or Anthropic, add it on the command-line like this: --llm MODEL_NAME
You will need to include an API key for the LLM. This can be OPENAI_API_KEY for an OpenAI LLM or ANTHROPIC_API_KEY for Anthropic.
You can also pass the API key to hespi with the --llm-api-key API_KEY argument.
More information on the command line arguments can be found in the Command Line Reference in the documentation.
There is another command line utility called hespi-tools which provides additional functionality.
See the documentation for more information.
To train the model with custom data, see the documention.
Robert Turnbull, Emily Fitzgerald, Karen Thompson, and Jo Birch from The University of Melbourne.

The paper describing the pipeline available in BioScience:
Turnbull, Robert, Emily Fitzgerald, Karen Thompson, and Joanne L. Birch. “Hespi: a Pipeline for Automatically Detecting Information from Herbarium Specimen Sheets.” BioScience (August 2025). DOI: 10.1093/biosci/biaf042.
You can also find the preprint of the paper on arXiv: arXiv:2410.08740.
Here is the BibTeX entry for the paper:
@article{hespi,
author = {Turnbull, Robert and Fitzgerald, Emily and Thompson, Karen M and Birch, Joanne L},
title = {Hespi: a pipeline for automatically detecting information from herbarium specimen sheets},
journal = {BioScience},
pages = {biaf042},
year = {2025},
month = {08},
abstract = {Specimen-associated biodiversity data are crucial for biological, environmental, and conservation sciences. A rate shift is needed to extract data from specimen images efficiently, moving beyond human-mediated transcription. We developed Hespi (for herbarium specimen sheet pipeline) using advanced computer vision techniques to extract authoritative data applicable for a range of research purposes from primary specimen labels on herbarium specimens. Hespi integrates two object detection models: one for detecting the components of the sheet and another for fields on the primary specimen label. It classifies labels as printed, typed, handwritten, or mixed and uses optical character recognition and handwritten text recognition for extraction. The text is then corrected against authoritative taxon databases and refined using a multimodal large language model. Hespi accurately detects and extracts text from specimen sheets across international herbaria, and its modular design allows users to train and integrate custom models.},
issn = {1525-3244},
doi = {10.1093/biosci/biaf042},
url = {https://doi.org/10.1093/biosci/biaf042},
eprint = {https://academic.oup.com/bioscience/advance-article-pdf/doi/10.1093/biosci/biaf042/63667847/biaf042.pdf},
}This research was supported by The University of Melbourne’s Research Computing Services and the Petascale Campus Initiative. The authors thank collaborators Niels Klazenga, Heroen Verbruggen, Nunzio Knerr, Noel Faux, Simon Mutch, Babak Shaban, Andrew Drinnan, Michael Bayly and Hannah Turnbull.
Plant reference data obtained from the Australian National Species List (auNSL), as of March 2024, using the:
- Australian Plant Name Index (APNI)
- Australian Bryophyte Name Index (AusMoss)
- Australian Fungi Name Index (AFNI)
- Australian Lichen Name Index (ALNI)
- Australian Algae Name Index (AANI)
and the World Flora Online Taxonomic Backbone v.2023.12, accessed 13 June 2024.
This pipeline depends on YOLOv8, torchapp, Microsoft's TrOCR.
Logo derived from artwork by ka reemov.
See the documentation for more information for references or use the command:
hespi-tools bibtex