This software is an experimental tool for Research Use Only. It is intended solely for research purposes related to the diagnosis of Developmental Dysplasia of the Hip (DDH). This software has NOT been approved as a medical device by any regulatory agency in any country. It is not intended for clinical use, diagnosis, or treatment. The software is provided "as is" without any warranty, express or implied. Use of this software is at your own risk. The developers and contributors are not responsible for any consequences resulting from the use or misuse of this software. Please consult with qualified healthcare professionals for any medical advice or diagnosis.
Retuve (from the scottish gaelic Ri taobh meaning beside) is a framework for analysing infant hips. It is designed to be a flexible and extensible framework that can be used by developers, AI researchers and clinicians.
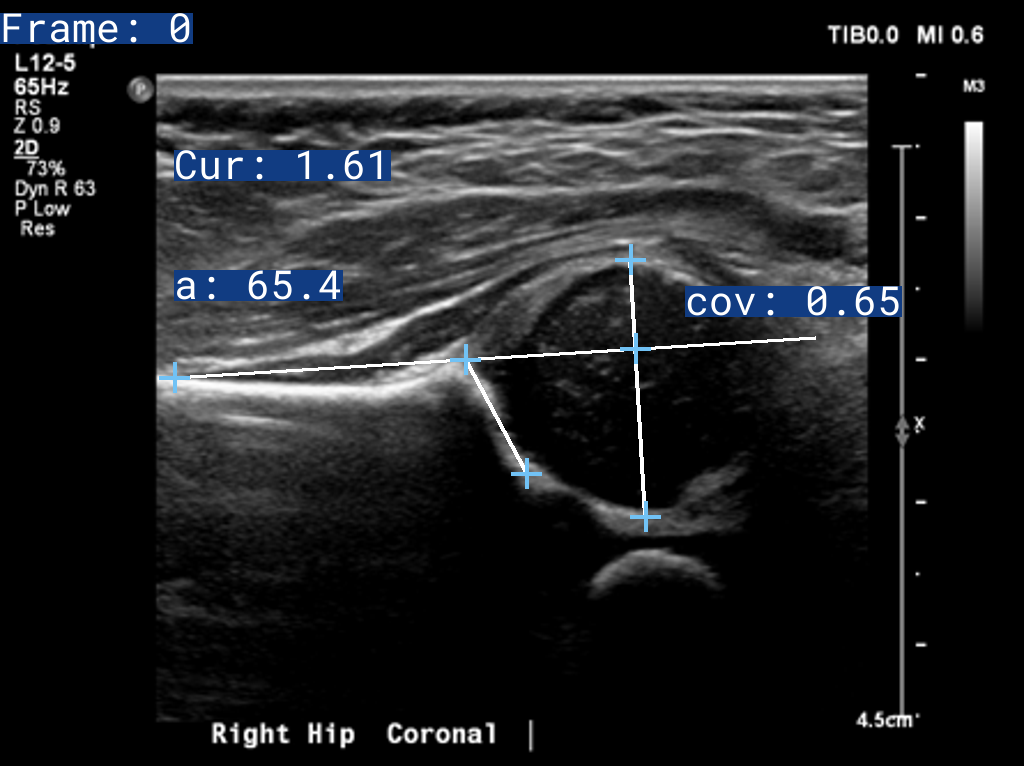
It takes in raw hip Ultrasound and X-Ray images, and outputs a report with the labelled images, and the results, exactly as a clinician would.
Attribution of the above Ultrasound Images: Case courtesy of Ryan Thibodeau from https://radiopaedia.org 172535 (https://radiopaedia.org/cases/172535)
Attribution of the above X-Ray Images: Fraiwan, Mohammad; Al-Kofahi, Noran; Hanatleh, Omar; ibnian, ali (2022), “A dataset of DDH x-ray images”, Mendeley Data, V2, doi: 10.17632/jf3pv98m9g.2
See our docs online at https://retuve.nidusai.ca
Retuve is licensed under the Apache 2.0 License. This means that you can use it for commercial purposes, and modify it as you see fit. The only requirement is that you must provide the license with any distribution of the software.
Please Note that the 2D Sweep and 3D DICOMs are sythetically stitched together from 2D Graf Hips, so do not expect accurate results. We are working on getting a 3DUS Case on a Creative Commons License to have as an example.
To get started with Retuve, you can install it via pip:
pip install git+https://github.com/radoss-org/retuve.gitYou can then run the following code to get a basic report:
WARNING: Before running this script, please make sure you have read the data disclaimer at DATA_DISCLAIMER.md.
import pydicom
from radstract.data.dicom import convert_dicom_to_images
from retuve.defaults.hip_configs import default_US
from retuve.defaults.manual_seg import manual_predict_us
from retuve.testdata import Cases, download_case
from retuve.funcs import analyse_hip_2DUS
# Example usage
dcm_file, seg_file = download_case(Cases.ULTRASOUND_DICOM)
dcm = pydicom.dcmread(dcm_file)
images = convert_dicom_to_images(dcm)
hip_data, img, dev_metrics = analyse_hip_2DUS(
images[0],
keyphrase=default_US,
modes_func=manual_predict_us,
modes_func_kwargs_dict={"seg": seg_file},
)
img.save("2dus.png")Attribution of the above Ultrasound Images: Case courtesy of Ryan Thibodeau from https://radiopaedia.org 172535 (https://radiopaedia.org/cases/172535)
- pip installable (easy to intergrate with you existing systems)
- Apache 2.0 Licensed
- AI is fully pluggable/modular
- Basic Web Interface bundled (through Retuve Trak)
- CLI Interface
- Swagger API Provided (through Retuve Trak)
Examples can be found at https://github.com/radoss-org/retuve/tree/main/examples
We have separate documentation for different use cases. Please see them all here:
We provide high level overviews for different types of users. This includes a tailored description of Retuve, and some highlighted features:
Retuve can analyse Hips for:
You can clone the repository and install the dependencies with the following command:
git clone https://github.com/radoss-org/retuve.gitYou can then install retuve with poetry, and then run the tests:
NOTE: These tests are about testing consistency between changes, and not directly testing the accuracy of the AI. See changenotes for tracking.
# Needed for the scripts
pip install poethepoet
cd retuve
poetry install
# Generate the test data
poe testgen
# Run all tests, including examples.
poe test_all
# Get info on all other dev scripts
poe helpWe are looking for support on the following:
- Landmark Models from litrature
- Native 3D Ultrasound Methods
We also accept any new algorithms for the different metrics, with the option to switch back to the old method provided through the config.
Please also see our Milestones for more information on what we are working on.
Data for the examples and tests is from https://github.com/radoss-org/radoss-creative-commons.
Please see this repo for attribution and licensing information.