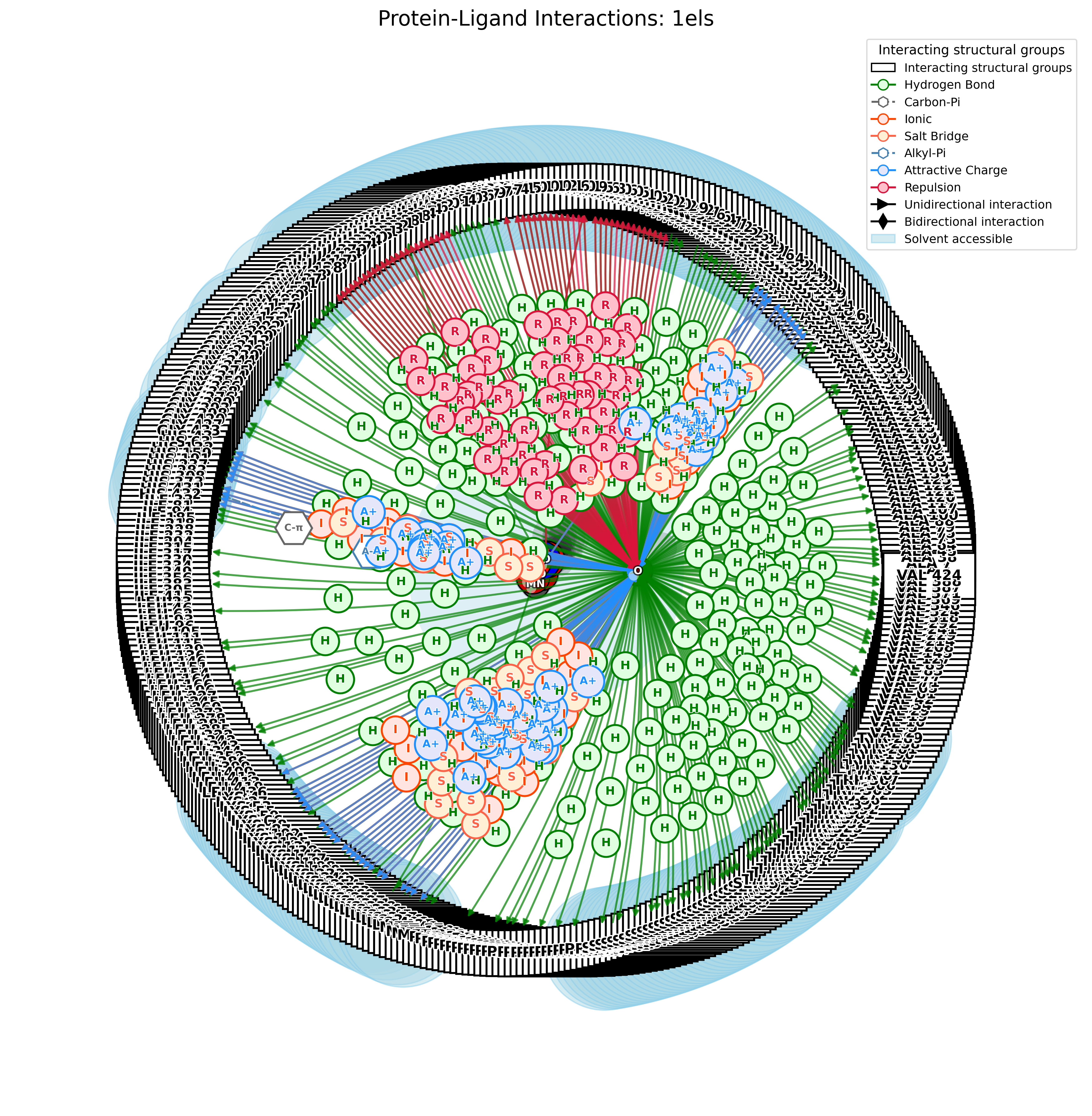
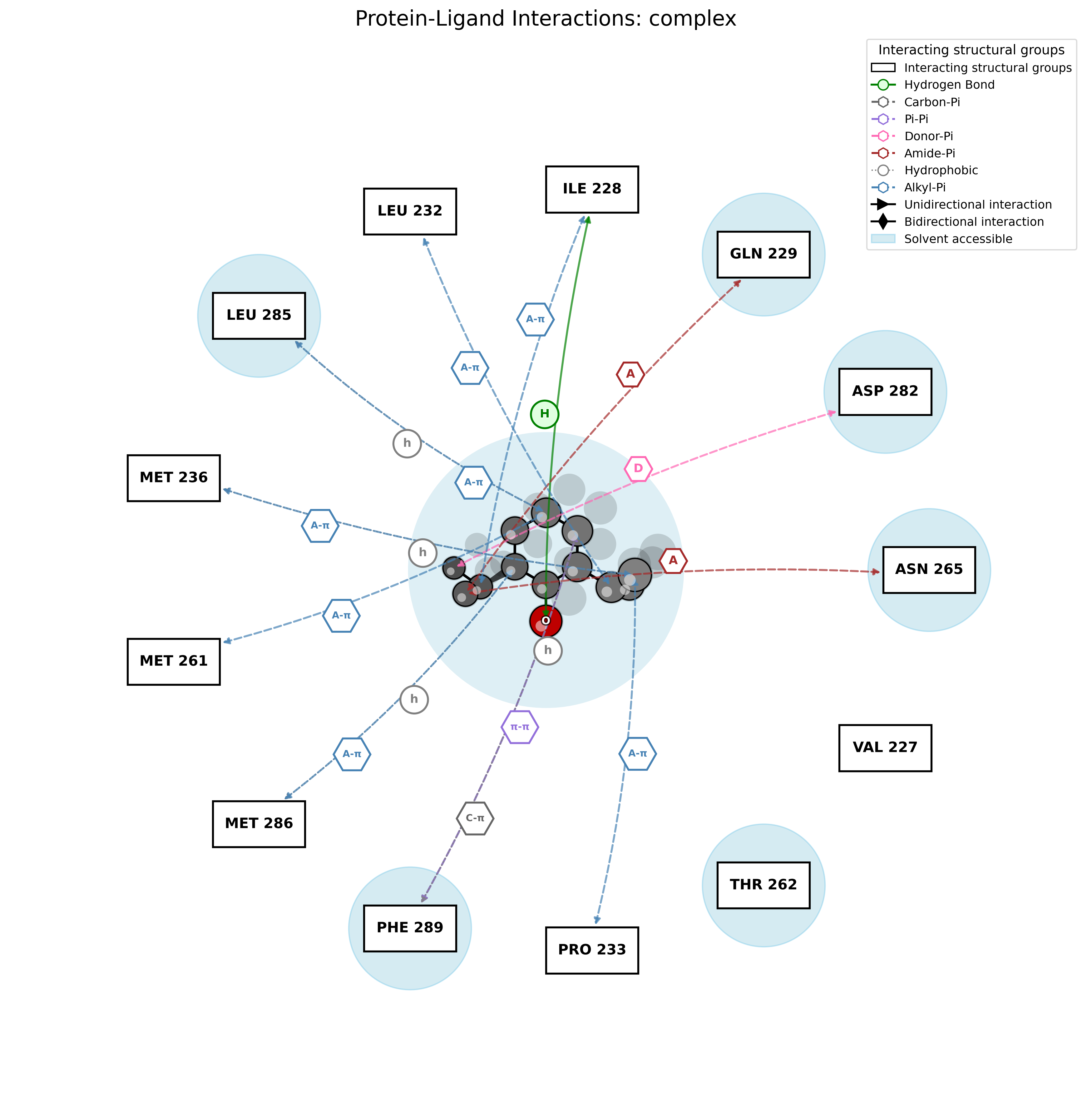
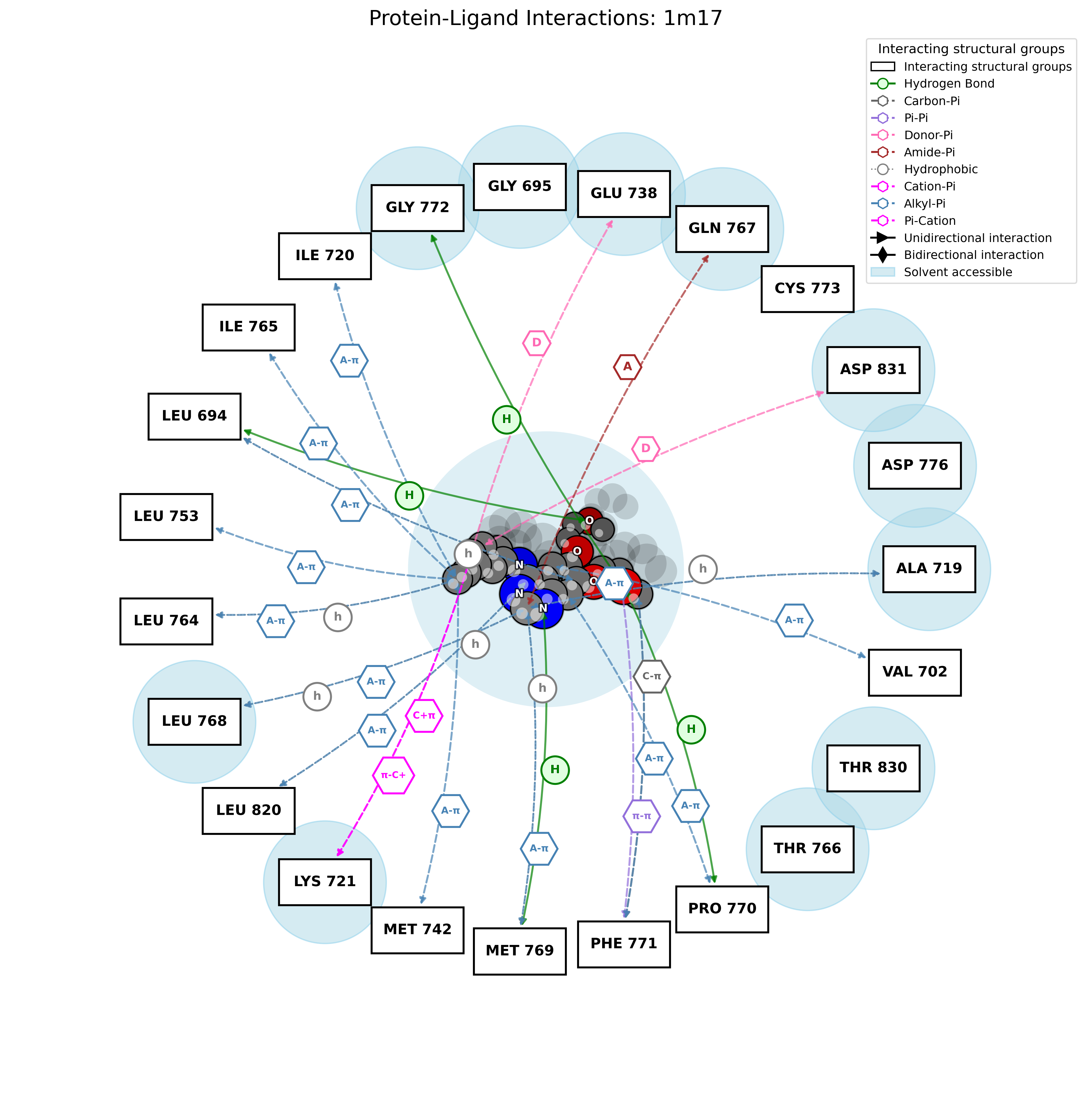
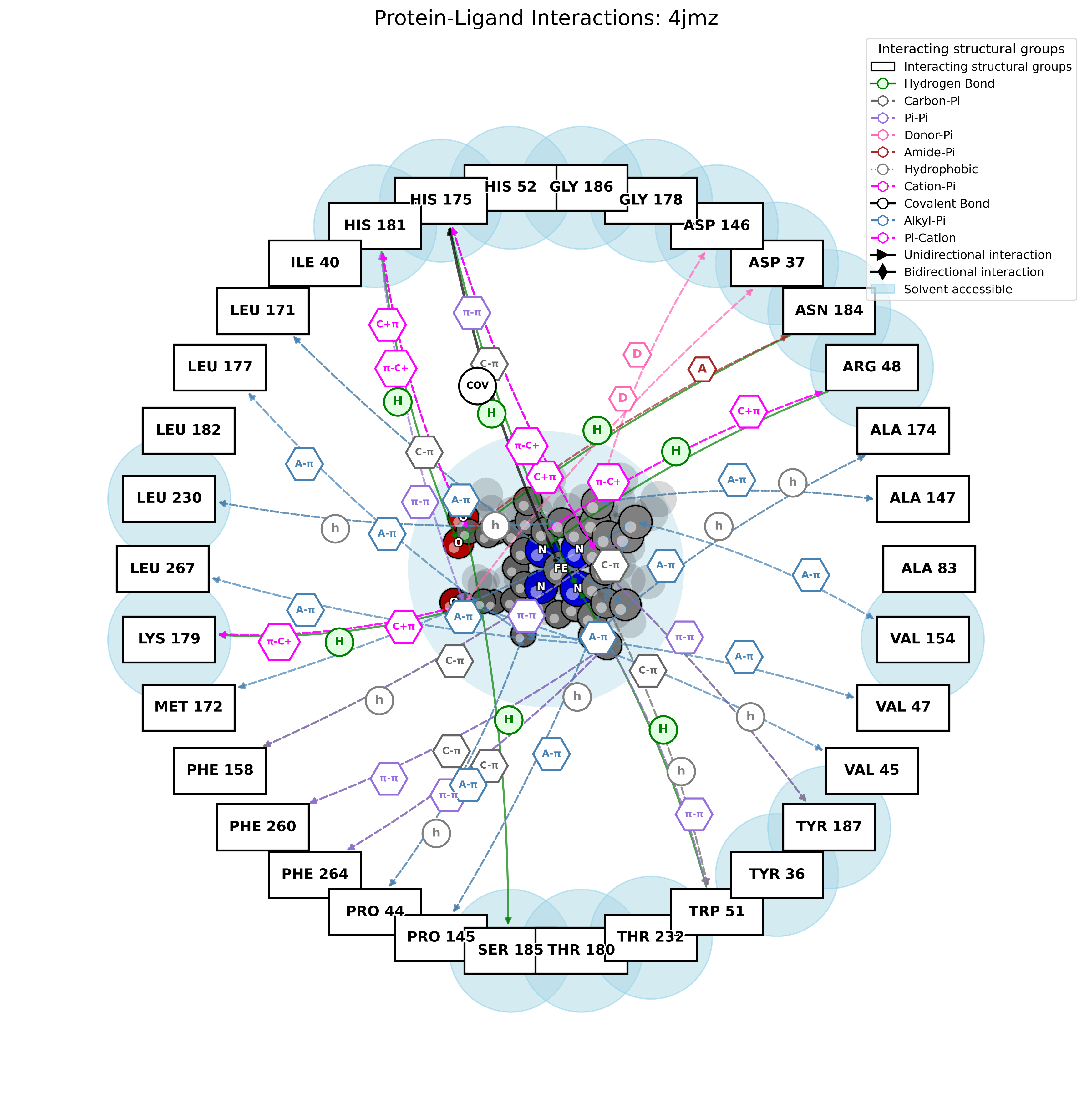
PandaMap: A Python package for visualizing protein-ligand interactions with 2D ligand structure representation.
Protein AND ligAnd interaction MAPper: A Python package for visualizing protein-ligand interactions with 2D ligand structure representation
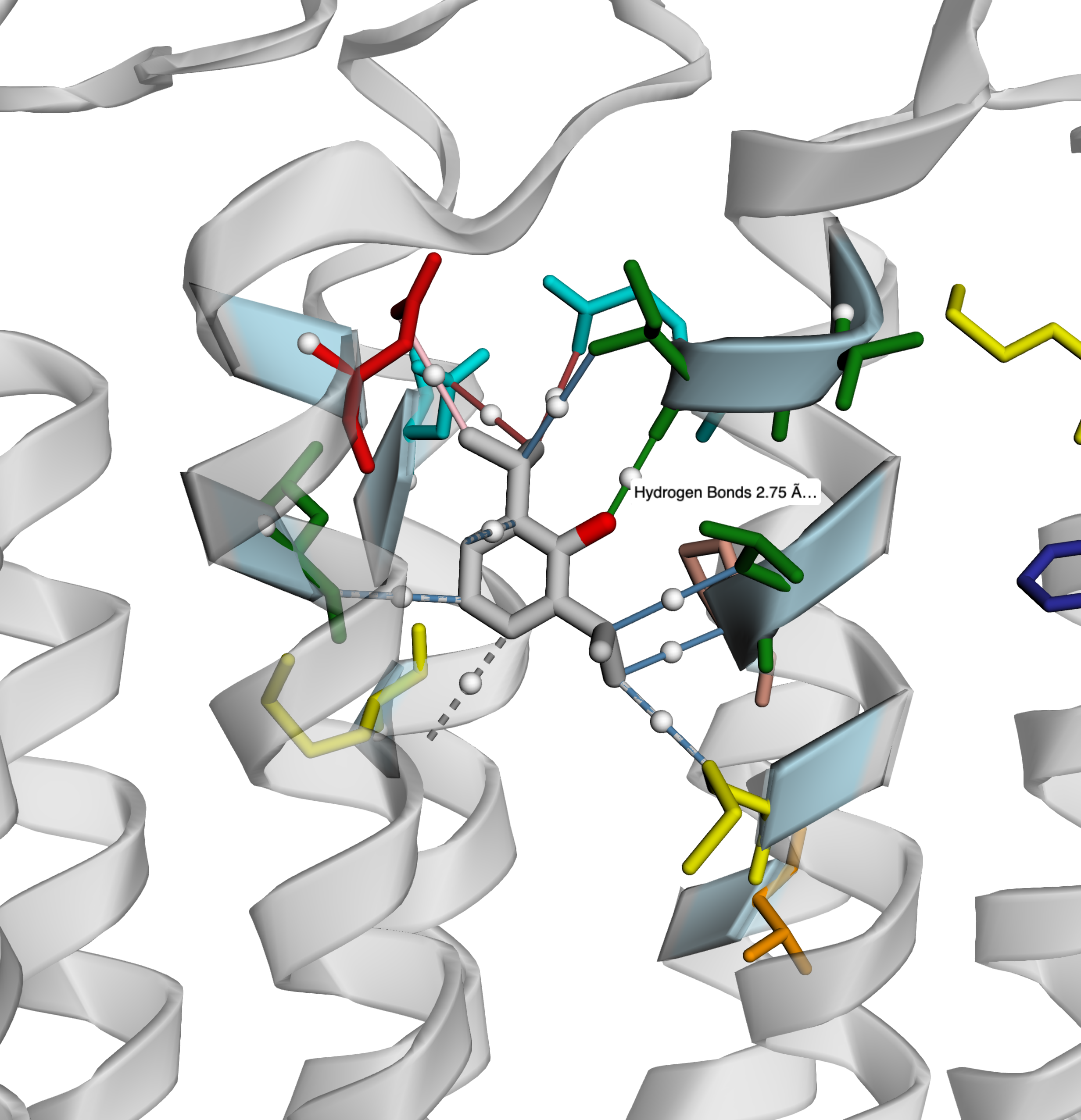
Version 4.1.0: With 3D HTML rendering
PandaMap is a Python package for visualizing protein-ligand interactions with enhanced detection methods.
- Visualization of protein-ligand complexes with 3D-enhanced 2D representations
- Comprehensive interaction detection:
- Hydrogen bonds
- Pi-Pi stacking
- Hydrophobic interactions
- Salt bridges and ionic interactions
- Halogen bonds
- Metal coordination
- And many more!
- Realistic solvent accessibility calculation
- Support for multiple input formats (PDB, CIF, PDBQT)
- Detailed interaction reports
- 3D Maps with rendering
👉 Click here for a live interactive 3D example
pip install pandamap-
Rich CLI Output (colored update messages):
pip install pandamap[fancy] -
3D HTML Visualization Support (via py3Dmol):
pip install pandamap[viz] -
All Features:
pip install pandamap[full]
- dssp #It can be installed externally
brew install dssp #mac users
sudo apt-get install dssp #linux users
Windows: Download from https://swift.cmbi.umcn.nl/gv/dssp/ - NumPy
- Matplotlib
- BioPython
pandamap -h
usage: pandamap [-h] [--output OUTPUT] [--ligand LIGAND] [--dpi DPI] [--title TITLE] [--version] [--report] [--report-file REPORT_FILE] [--3d] [--3d-output OUTPUT_3D] [--no-surface] [--width WIDTH] [--height HEIGHT] [--no-3d-cues] structure_file
PandaMap: Visualize protein-ligand interactions from structure files
positional arguments:
structure_file Path to structure file (PDB, mmCIF/CIF, or PDBQT format)
options:
-h, --help show this help message and exit
--output, -o OUTPUT Output image file path
--ligand, -l LIGAND Specific ligand residue name to analyze
--dpi DPI Image resolution (default: 300 dpi)
--title, -t TITLE Custom title for the visualization
--version, -v Show version information
--report, -r Generate text report
--report-file REPORT_FILE
Output file for the text report (default: based on structure filename)
--3d Generate an interactive 3D visualization of protein-ligand interactions
--3d-output OUTPUT_3D
Output file path for 3D visualization (default: based on input filename)
--no-surface Do not show protein surface in 3D visualization
--width WIDTH Width of 3D visualization in pixels (default: 800)
--height HEIGHT Height of 3D visualization in pixels (default: 600)
--no-3d-cues Disable 3D cues in 2D visualization
## 🐼 PandaMap — Protein AND ligAnd interaction MAPper
Visualize protein-ligand interactions in both **2D image format** and **interactive 3D HTML** with optional reports.
---
### 🚀 Command-Line Usage
```bash
# Basic 2D interaction map
pandamap protein_ligand.pdb --output interactions.png
# With specific ligand
pandamap complex.cif --ligand LIG --output cif_interaction.png
# Generate text report
pandamap 4jmz.pdb --ligand HEM --report --report-file HEM.txt
pandamap 1m17.pdb --ligand AQ4 --report --report-file 1m17.txt
# Full analysis: image + report
pandamap complex.pdb --ligand PFL --output complex.png --report --report-file complex.txt# Basic 3D HTML map
pandamap protein_ligand.pdb --3d
# Save HTML to custom file
pandamap complex.pdb --ligand LIG --3d --3d-output interaction.html
# Full combo: 2D image + 3D HTML + report
pandamap structure.pdb --ligand LIG --output map.png --3d --3d-output map.html --report --report-file map.txt
# Customize 3D size
pandamap structure.pdb --3d --width 1024 --height 768 --no-surfacefrom pandamap import HybridProtLigMapper
# Basic initialization
mapper = HybridProtLigMapper("protein_ligand.pdb", ligand_resname="LIG")
# Run full workflow and generate 2D image
mapper.run_analysis()
mapper.visualize(output_file="interactions.png")
# Generate interactive 3D HTML
from visualization_3d import create_pandamap_3d_viz
create_pandamap_3d_viz(mapper, output_file="interactions.html")# Use DSSP for accurate surface exposure detection
mapper.run_analysis(use_dssp=True)
mapper.visualize(output_file="with_dssp.png")Or fallback to Python-only:
mapper.run_analysis(use_dssp=False)
mapper.visualize(output_file="pure_python.png")from improved_interaction_detection import ImprovedInteractionDetection
mapper = HybridProtLigMapper("protein.pdb", ligand_resname="LIG")
mapper.run_analysis()
# Optional: refine results
detector = ImprovedInteractionDetection()
filtered = detector.refine_interactions(mapper.interactions)
# Write to report
detector.generate_report(
ligand_metadata={
'hetid': mapper.ligand_residue.resname,
'chain': mapper.ligand_residue.parent.id,
'position': mapper.ligand_residue.id[1],
'longname': mapper.ligand_residue.resname,
'type': 'LIGAND',
},
interaction_data=filtered,
output_file="filtered_report.txt"
)pandamap -h
pandamap --version=============================================================================
PandaMap Interaction Report
=============================================================================
Ligand: PAH:A:439
Name: PAH
Type: LIGAND
------------------------------
Interacting Chains: A
Interacting Residues: 13
------------------------------
Interaction Summary:
Hydrogen Bonds: 10
Carbon-π Interactions: 1
Metal Coordination: 4
Ionic Interactions: 2
Salt Bridges: 2
Alkyl-π Interactions: 1
Attractive Charge: 2
Repulsion: 5
------------------------------
Hydrogen Bonds:
1. GLU168A -- 2.66Å -- PAH
2. ASP246A -- 2.60Å -- PAH
3. GLN167A -- 3.10Å -- PAH
4. ASP320A -- 3.46Å -- PAH
5. LYS396A -- 3.05Å -- PAH
6. SER375A -- 2.82Å -- PAH
7. SER39A -- 3.06Å -- PAH
8. ARG374A -- 2.98Å -- PAH
9. GLY37A -- 3.36Å -- PAH
10. LYS345A -- 3.21Å -- PAH
------------------------------
Carbon-π Interactions:
1. HIS373A -- 4.29Å -- PAH
------------------------------
Metal Coordination:
1. ASP246A -- 2.24Å -- PAH
2. GLU295A -- 2.24Å -- PAH
3. ASP320A -- 2.19Å -- PAH
4. GLY37A -- 2.12Å -- PAH
------------------------------
Ionic Interactions:
1. ARG374A -- 2.98Å -- PAH
2. LYS345A -- 3.21Å -- PAH
------------------------------
Salt Bridges:
1. ARG374A -- 2.98Å -- PAH
2. LYS345A -- 3.21Å -- PAH
------------------------------
Alkyl-π Interactions:
1. HIS373A -- 5.27Å -- PAH
------------------------------
Attractive Charge:
1. ARG374A -- 2.98Å -- PAH
2. LYS345A -- 3.21Å -- PAH
------------------------------
Repulsion:
1. ASP320A -- 3.46Å -- PAH
2. GLU168A -- 3.10Å -- PAH
3. ASP246A -- 2.60Å -- PAH
4. GLU295A -- 3.95Å -- PAH
5. GLU211A -- 4.34Å -- PAH
=============================================================================
If you use PandaMap in your research, please cite:
Pritam Kumar Panda. (2025). Protein AND ligAnd interaction MAPper: A Python package for visualizing protein-ligand interactions with 2D ligand structure representation. GitHub repository. https://github.com/pritampanda15/PandaMap
This project is licensed under the MIT License - see the LICENSE file for details.