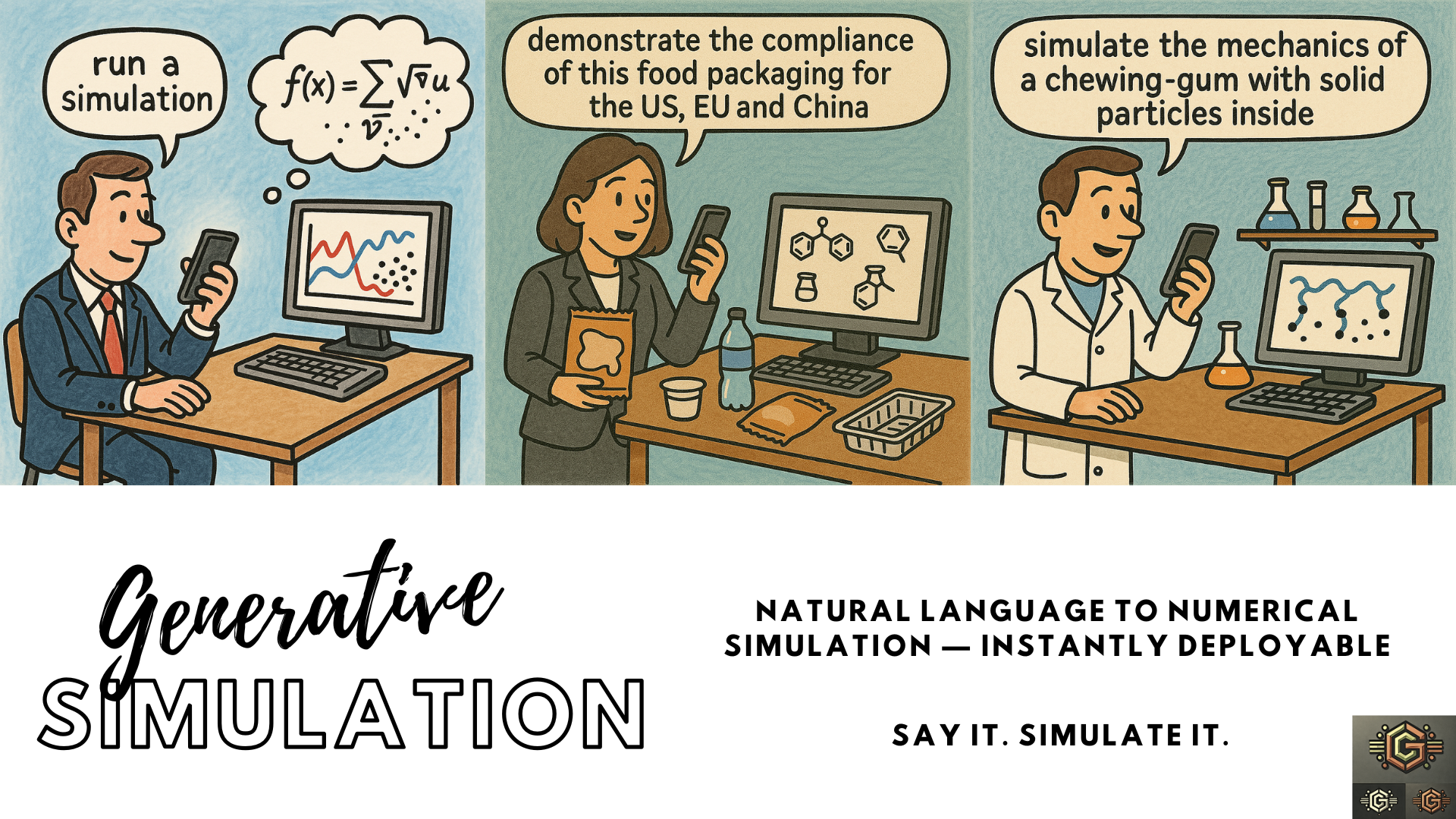
Generative Simulation (GS) is an emerging paradigm that develops advanced numerical simulations using elemental "bricks" accessible to text-generative AI models and GS-agents. Learn more on GS-agents on GS-Hub project.
🧠 Tools like ChatGPT and its consorts excel at programming across many languages and grasping high-level, macroscopic concepts. However, they cannot seamlessly connect physics, chemistry, biology, and mathematics to solve real-world scientific and engineering problems.
Generative Simulation provides modular bricks—representations of physical or conceptual building blocks—that Large Language Models (LLMs) can understand and manipulate to bridge this gap. These bricks can be designed with AI assistance, but the overarching logic, scientific insight, and problem-specific nuance remain in human hands.
Once the bricks form a structured language, the subsequent stages of model development, simulation assembly, or code generation can be delegated back to the AI and iteratively refined under human supervision.
🧩 These bricks provide a clear context that LLMs can follow within their existing context window (from 8K to 128K tokens). Simulations and scenarios can be produced from prompts including specific instructions and experimental data (e.g., chromatograms). Clear examples, reusable classes, and operators overcome the limitations of the current window size.
🚧 A prototype demonstrating LFCL orchestration is under development and is drafted in this document.
🎨 Credits: Olivier Vitrac
- 📦 Some Showcases
- 🍕 Example 1 | Pizza³ – A Multiscale Python Toolkit
- 🍽️ Example 2 | SFPPy – Compliance & Risk Assessment for Food Contact Materials
- 🌐 Example 3 | SFPPylite – SFPPy in Your Browser
- 🧪⚛️ Example 4 | Radigen – Kernel for Radical Oxidation Mechanisms
- 📡🧬 Example 5 | sig2dna – Symbolic Fingerprinting of Analytical Signals
- 🧪 Part 1 | Language-First Computational Lab (LFCL) Road Map
- 🧰 Part 2 | LFCL Architecture (Sketch)
- 🧩🌱 Part 3 | Building Specialized Kernels
- 🤖 Part 4 | GS-Agents & the GSHub Initiative
- 👥👉 Want to contribute?
🔧 The Generative Simulation Ecosystem already includes powerful computational and multiscale kernels for:
-
⚙️ Mechanics – particle-based models and forcefields (LAMMPS-compatible): large deformations, rupture, adhesion/melting
-
💧 Mass Transfer – diffusion–reaction systems (Fickian PDEs, sorption models, molecular diffusivity estimators)
-
🔥 Chemical Reactivity – ODE networks, combinatorial pathway generators, graph-based transformations
-
📡 Chemical Signal Encoding and Analysis – symbolic transformation of 1D analytical signals (GC-MS, NMR, FTIR…)
📚 Each kernel is enriched with embedded domain-specific knowledge and scientific databases:
- 🧪 Forcefields — atomistic, coarse-grained (DPD, SDPD, SPH)
- 🧊 Thermodynamic Data — solubilities, mixing energies, phase transitions
- 🧬 Transport Properties — diffusivity, dissolution, activation energy models (Arrhenius, free-volume)
- ⚗️ Reactivity Schemes — reaction networks, rate laws, temperature effects
- ⌬ Chemical Databases — PubChem integration, CAS handling, identifiers
- ⛓️ Material Databases — polymer properties (e.g.,
$T_g$ , density, crystallinity)- ☢️ Toxicology Tools — ToxTree predictions, hazard flags
- ⚖️ Regulatory Frameworks — legal thresholds (🇪🇺 EU, 🇺🇸 US, 🇨🇳 China)
- Chemical Fingerprints — symbolic encoding of GC-MS, FTIR, RAMAN, $^1$H-NMR
💡 Many models also integrate practical factors across the value chain:
- ♻️ Recycling & decontamination processes for plastics and cellulosic materials
- 🧃🥡 Packaging types, geometries, and market statistics
- 🥗🍛 Food matrix composition and reactivity
- ♨️🌡️❄️ Food transformation (e.g., deep-frying, pasteurization) and storage
🧠 All components are designed as modular Python objects readable by LLMs, enabling simulation-driven reasoning, scenario exploration, and code generation through natural language prompts.
Examples Overview
Pizza3 simulates the mechanics of soft matter using LAMMPS, with fully reusable Python objects that AI can understand and extend. This modular approach simplifies multiscale physics integration while abstracting away unnecessary technicalities for HPC/cloud deployment.
- 💻 Computational resources: ++++
- 🧠 Complexity: +++
- 🚀 Typical deployment: HPC, GPU, AWS
- 🔗 Source: Pizza3 on GitHub
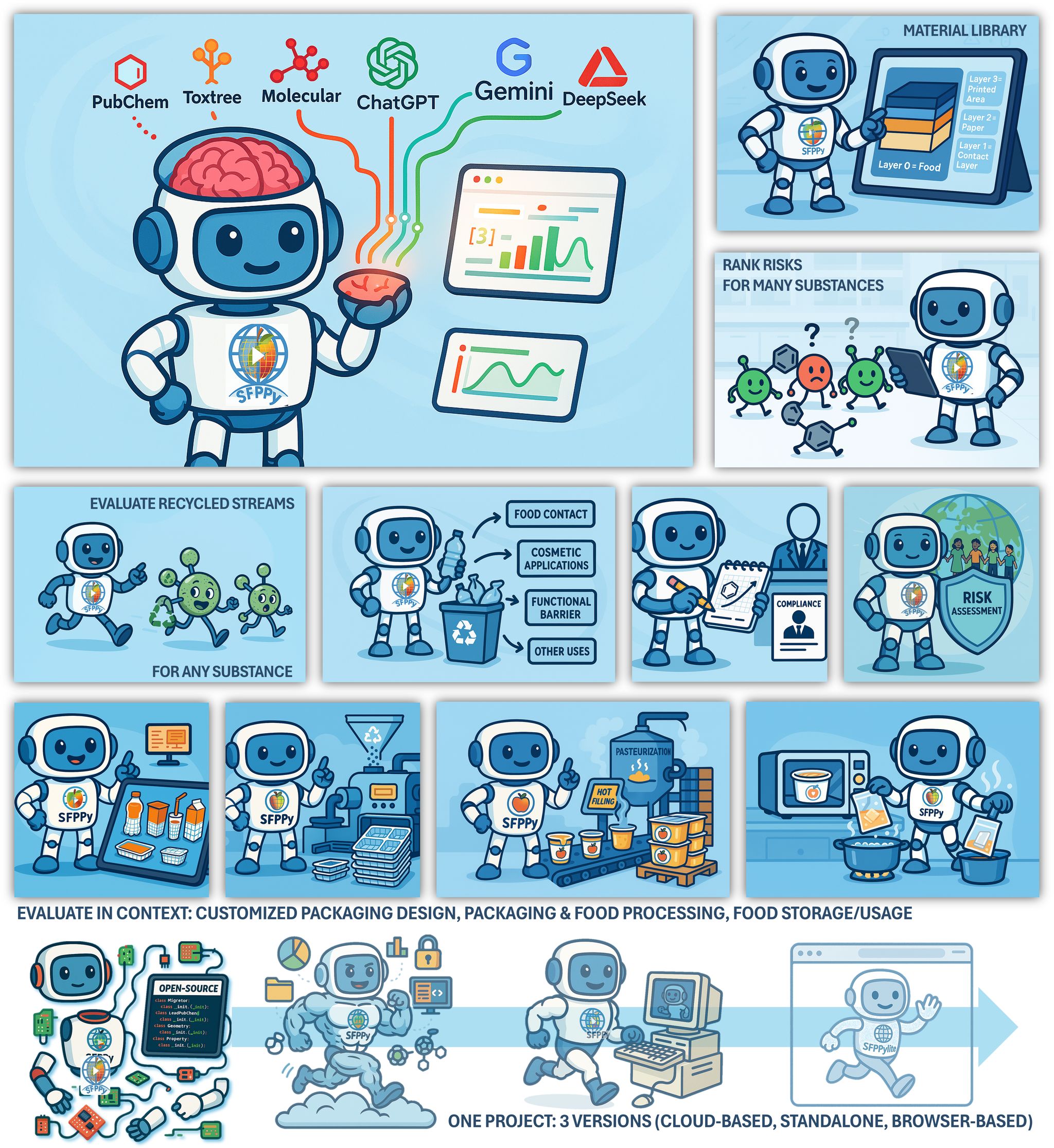
SFPPy is a Python framework that accelerates the evaluation of material safety in contact with food. It connects to regulatory databases, computes migration properties from molecular structures, and simulates mass transfer across packaging layers.
Already recognized by EU, US, and Chinese agencies, SFPPy integrates:
-
Mass transfer models
-
AI-generated scenarios
-
Jupyter-based documentation and reports
-
💻 Computational resources: +/++
-
🧠 Complexity: ++
-
🖥️ Deployment: Colab, Jupyter Server
-
📘 Usage: Notebooks for modeling, tracking, and AI-assisted reporting
-
🔗 Source: SFPPy on GitHub
SFPPylite is a WebAssembly-based version of SFPPy running entirely in your browser—no server, no installation required.
Perfect for:
-
🏫 Classroom training
-
🧪 SME simulations
-
📝 Regulatory collaboration (copy/paste into chatbots or notebooks)
-
💻 Computational resources: +
-
🧠 Complexity: ++
-
🖥️ Deployment: Browser (Chrome, Firefox)
-
🔗 Source: SFPPylite on GitHub
Radigen simulates radical oxidation in edible oils, biofuels, and polymer degradation. It uses reactive functions, class inheritance, and mechanistic logic to build large-scale reaction networks with thousands of time-dependent species.
Composable Python bricks:
mixturedefines species and reactionsmixtureKineticssolves the ODE systemlumpedaggregates similar speciesTKO2cyclecontrols T/O₂ cycles
💬 Prompt example:
“Simulate oxidation of linoleic acid for 3 days at 160°C and partial renewal with fresh oil.”
- 💻 Computational resources: +
- 🧠 Complexity: +/++
- 🖥️ Deployment: Scripts, notebooks, LLM-assisted workflows
- 🔬 Usage: Food, pharma, cosmetics, polymer stability
- 🔗 Source: Radigen on GitHub
sig2dna transforms complex analytical signals—like GC-MS, NMR, or FTIR outputs—into compact, symbolic DNA-like sequences using a multi-scale wavelet encoding approach. These sequences enable motif recognition, signal alignment, and unsupervised classification, unlocking new capabilities for AI-assisted analysis of real-world chemical data.
This symbolic transformation compresses large signals (>95%) while preserving morphological detail. Once encoded, symbolic distances (e.g., Excess Entropy, Jaccard, Levenshtein, Jensen-Shannon) allow clustering, outlier detection, and blind source separation, even for overlapping or noisy spectra.
sig2dna is especially suited for:
- ♻️ NIAS (non-intentionally added substances) fingerprinting in recycled materials
- 🧪 Quality control in chemical, food, or cosmetic analysis
- 🤖 LLM-based workflows that search or cluster chemical patterns from symbolic codes
💬 Prompt example: “Classify this mixture of GC-MS signals using entropy distance and extract all motifs matching
YAZB.”
-
💻 Computational resources: +
-
🧠 Complexity: ++
-
🖥️ Deployment: Python scripts, Colab, Jupyter
-
📊 Usage: Signal classification, clustering, fingerprinting, AI-assisted interpretation
-
🔗 Source: sig2dna on GitHub
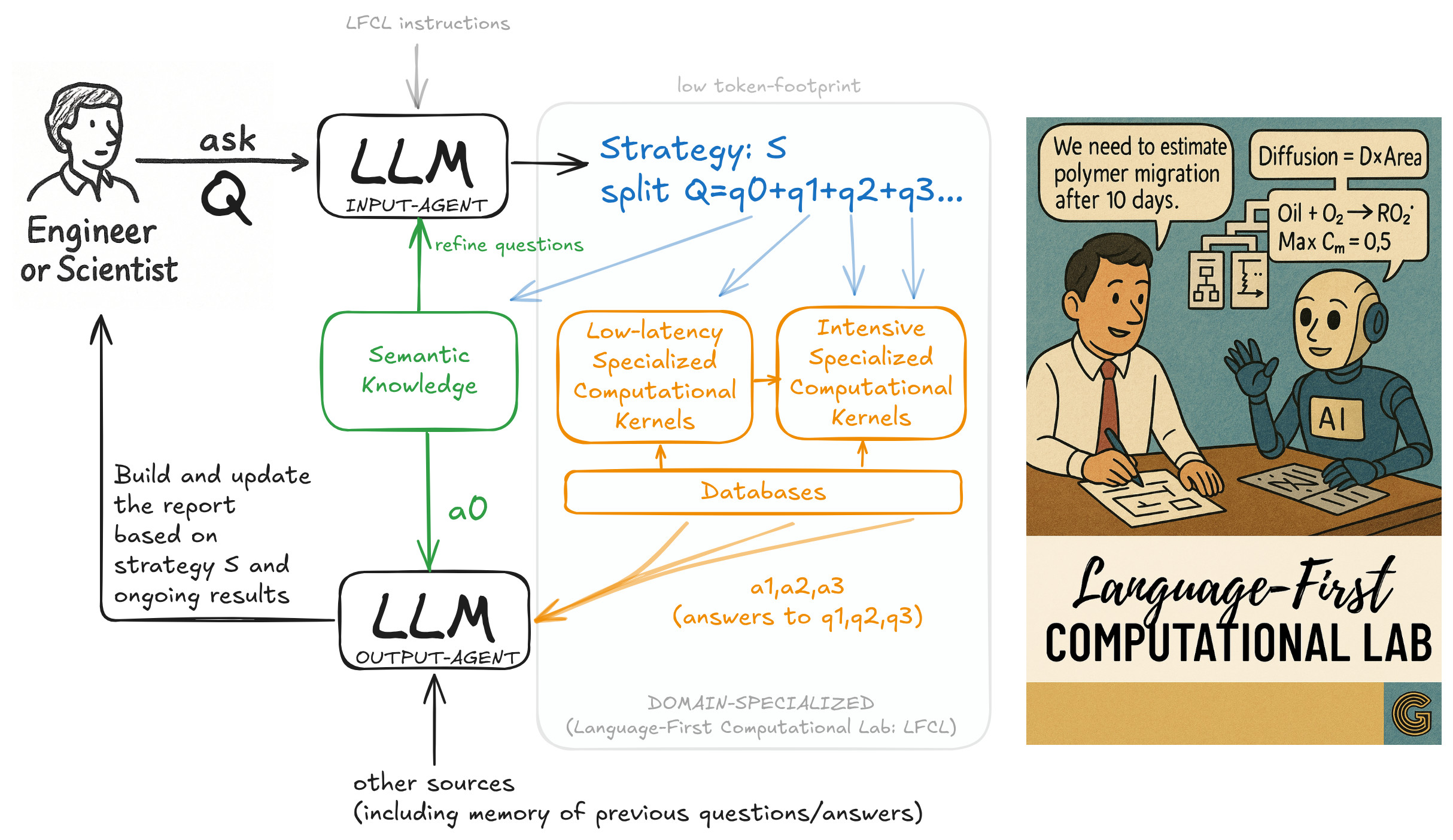
Generative Simulation enables a Language-First Computational Lab (LFCL)—a new paradigm where LLMs orchestrate numerical simulations using modular physics/chemistry/biology kernels with human-guided workflows.
In a language-first lab, natural language becomes the primary interface for composing, querying, refining, and explaining scientific models.
LLMs:
- 🛠️ Do not hold full models
- 🤖 Control, audit, and refine simulations via external kernels
- 🧠 Provide bidirectional reasoning with humans
Modular simulation kernels accessible via APIs, ideally stateless:
- Input: structured physical quantities
- Output: machine-readable + human-readable
Examples:
mass_transfer_kernel(diffusivity, geometry, time)radigen_kernel(reaction_network, T, dt)
LLM capabilities include:
- Interpretation (e.g., "what if T ↑ 10°C?")
- Scripting DSL or Python code
- Fitting, analysis, assumption validation
- Detecting uncertainty or ill-posed logic
Buffers tailored to 8K–128K token windows:
- Tracks assumptions, results
- Summarizes intermediate states
- Enables iterative refinement
Dashboards or notebooks to:
- Override model logic
- Display results/uncertainties
- Compare and version simulations
graph TD
A[User Input<br>Natural Language Prompt]
A -->|interpreted| B[<b>LLM Agent</b]
B -->|calls| C[Simulation Kernel]
C --> D[Output: Structured + Natural Language]
D -->|interpreted| B
B --> E[Notebook / Dashboard]
E --> A
Humans (engineers, scientists, technicians) define and guard simulation logic:
- 🎯 Define kernel schemas
- 🔍 Validate logic
- 📐 Audit assumptions
- 🔁 Design prompt workflows
The human becomes the workflow architect.
The applications are numerous. Current kernels can already provide answers in several domains.
| Domain | Example Workflow |
|---|---|
| Polymer Migration | “Estimate PET monomers migration after 10 days at 60°C” |
| Oxidation Kinetics | “Which species peaks first during FAME oxidation at 90°C?” |
| Thermomechanics | “Simulate stress in packaging during sterilization” |
| Circular Design | “Propose recycled polymer blends with acceptable safety and strength” |
| Haptic and Mouthfeel perception | “Simulate the texture perception of a mixture of ….” |
To scale beyond context limits:
- Use external caches or memory modules
- Break work into episodes with symbolic anchors
- Export results as summarized memory tokens
The LFCL architecture enables an engineer or scientist to delegate complex scientific questions to an LLM-powered orchestration system. Upon receiving a question Q, the LLM Input-Agent devises a strategy S that decomposes Q into sub-questions (q₀, q₁, q₂, ...), each routed to an appropriate computational process:
- q₀: interpreted through semantic knowledge
- q₁, q₂, ...: dispatched to low-latency or intensive specialized kernels, drawing on numerical databases The answers a₀, a₁, a₂, ... are consolidated by the LLM Output-Agent, which builds a structured response and report.
🎨 Credits: Olivier Vitrac
This architecture ensures reasoning and simulation are balanced, reducing delays and computational cost while preserving scientific rigor and traceability.
This architecture supports real-time orchestration of physical/chemical/biological kernels by LLMs using token-efficient prompts.
graph TD
A["👩🔬 USER<br>(Scientist / Engineer)"]
A --> B["🗣️ Natural Language Prompt"]
B --> C["🧠 <b>LLM Interpreter</b><br>(Language Layer)"]
C --> D["🧾 Function Call / Code Generation"]
D --> E["⚙️ Simulation Kernels<br>(Physics / Chemistry / Biology)"]
E --> F["📊 Results<br>(Structured Data)"]
F --> G["🔁 <b>LLM Interpreter</b><br>(Backloop)"]
G --> H["📝 Narrative Summary + Scientific Explanation"]
LFCL/
├── main.py # Entry point
├── agent.py # LLM orchestrator
├── memory.py # Token-aware memory manager
├── interface.py # CLI, notebook or Streamlit
├── kernels/
│ ├── radigen_interface.py # Oxidation modeling API
│ ├── sfppy_interface.py # Migration modeling API
│ └── core_utils.py # Shared utilities
├── prompts/
│ └── templates.md # Prompt interpreters
├── config/
│ └── kernel_schema.yaml # I/O standards
└── logs/
└── session_001.jsondef run_oxidation_simulation(species_list, T=80, duration=("10", "d")):
from radigen.radigen3.mixtureKinetics import mixtureKinetics
mk = mixtureKinetics.from_species(species_list)
mk.solve(time=duration, T=T)
return mk.get_dataframe()Minimal API:
from_species(...).solve(...).get_dataframe()
def handle_prompt(prompt, context):
if "oxidation" in prompt and "FAME" in prompt:
species = ["monoAllylicCOOH", "oxygen"]
T = 90
time = (10, "h")
from kernels.radigen_interface import run_oxidation_simulation
df = run_oxidation_simulation(species, T, time)
return summarize_results(df, species)class ContextMemory:
def __init__(self, max_tokens=8000):
self.entries = []
self.max_tokens = max_tokens
def add(self, text):
self.entries.append(text)
self._trim()
def _trim(self):
...Tracks:
- Results
- Assumptions
- Code chunks
import streamlit as st
from agent import handle_prompt
prompt = st.text_area("Ask your lab:", "Simulate oxidation of FAME at 90°C")
if st.button("Run"):
result = handle_prompt(prompt, context={})
st.write(result)# Template: Oxidation Modeling
**User Prompt**
"Can you simulate the oxidation of methyl linoleate at 80°C for 5 days?"
**Parsed Actions**
- domain = oxidation
- model = radigen
- species = ["monoAllylicCOOH", "oxygen"]
- time = (5, "d")
- T = 80°C
**Kernel Call**
run_oxidation_simulation(...)
**Summary**
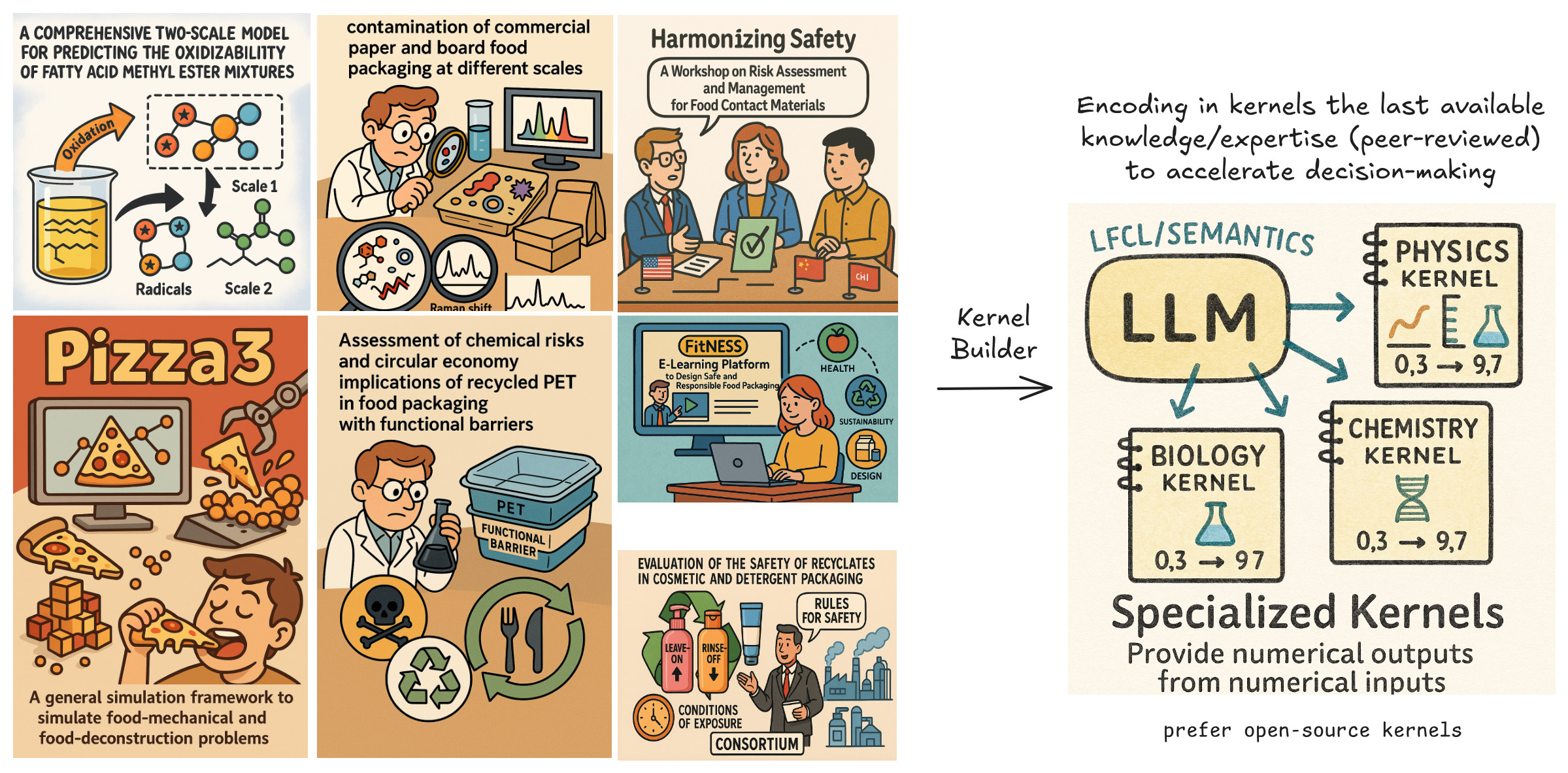
Simulated methyl linoleate oxidation. Products include L1OOH, ROO•. See plots.Specialized kernels are domain-specific computational modules that extend the reasoning capabilities of language models into the realm of quantitative science and engineering. They are the physical, chemical, or biological “brains” of the Language-First Computational Lab (LFCL)—enabling LLMs to compute, not just communicate.
Unlike general-purpose LLMs, which rely on linguistic priors, these kernels:
- Accept numerical inputs (e.g., lab data, physical parameters, boundary conditions)
- Deliver quantitative outputs (e.g., reaction kinetics, migration rates, stress/strain curves)
- Provide high precision, traceability, and interpretability far beyond generic AI reasoning
🧠 Through the LFCL interface, these kernels are made accessible to LLMs via semantic hooks and structured prompts—allowing agents to reason over data, not just text.
🎨 Credits: Olivier Vitrac
- Each kernel must implement a LFCL-compatible interface, ideally in Python, to support both LLM integration and human usage.
- Kernel development should be driven by existing validated scientific methods—those supported by peer-reviewed literature, regulatory bodies, or industrial standards.
- Clear usage documentation is mandatory, including:
- Valid domains of application
- Acceptable input/output ranges
- Explicit warnings for extrapolation or domain gaps
- Associated uncertainty quantification
- The kernel type (⚡ low-latency vs 🧮 intensive) should match:
- The trustworthiness of the method
- The urgency or tier of the decision-making process
- The available computational resources
- Wherever possible, multitiered or progressive strategies should be used—starting with fast approximations and refining only where necessary.
- Kernels are most powerful when combined across domains (e.g., mass transfer + toxicology, or diffusion + reaction mechanics).
- LFCL supports both rough estimators and high-fidelity solvers (e.g., particle-based, mesh-based, Monte Carlo, or thermodynamic integration).
- Registries should list:
- Method provenance (e.g., peer-reviewed, regulatory, internal R&D)
- Maintenance status and contributors
- Example use cases and known limitations
- Open repositories are encouraged to foster independent validation, reusability, and collective improvement beyond the LLM ecosystem.
LLM agents can chain together past requests and kernel invocations to form new composite simulations. This enables deeper scenario construction and fosters creative reuses of existing scientific bricks.
In essence, LFCL provides augmented scientific intelligence for both expert and non-expert users—amplifying exploration and guiding attention toward meaningful results. It acts as a catalyst for more robust reasoning, faster hypothesis testing, and greater transparency.
SFPPy is a fully integrated kernel designed for food contact compliance and risk assessment. It combines:
- A robust mass transfer solver
- Molecular and toxicological databases
- Structure-based migration models
- A comprehensive documentation layer
- Strong alignment with EU, US, and Chinese regulatory frameworks
🎨 Credits: Olivier Vitrac
As a kernel in the LFCL ecosystem, SFPPy empowers LLMs to interpret and solve complex regulatory and technological queries in natural language—spanning:
- Material design & formulation
- Food type & processing conditions
- Packaging structure & multi-layer barriers
- Migration limits & consumer exposure scenarios
⏱️ What would traditionally take hours of expert interpretation can now be framed, explored, and drafted in seconds, with results that are explainable, traceable, and reproducible.
Example query: "Can you demonstrate that this recycled multilayer material complies with food contact regulations for fatty foods in the EU market after retort processing?"
Thanks to its structured design and regulatory foundations, SFPPy can simulate such scenarios directly or support them with partial answers, identifying gaps or missing inputs.
As Generative Simulation evolves, it becomes increasingly important to bridge natural language interaction with composable scientific kernels. GS-Agents are intelligent interfaces designed to do exactly that: they interpret scientific questions, invoke simulation kernels, and archive their reasoning paths.
The companion project, GSHub, provides the open infrastructure for these agents. It includes:
- A registry of interoperable kernels (e.g.,
radigen,SFPPy,sig2dna) - A memory system for storing prompt–response–result chains
- Tools for problem submission, review, and collaborative refinement
📬 Visit GSHub to explore how GS-Agents create a persistent, modular, and evolving form of machine–human reasoning. Contributions and forks welcome.
📌 Want to contribute? Reach out via GitHub issues or discussions to propose new kernels, interface improvements, or industrial/educational use cases!
🎨 Credits: Olivier Vitrac
olivier.vitrac@gmail.com | May 2025