This repository was built as a small demonstration of my coding skills, data management and applied knowledge of deep learning techniques. The selected task is derived from the Luna16 challenge. The task is to classify whether clinical follow-up is required, based on the provided coordinates of a specific lung nodule candidate. Due to the limited time which was available to me, the intended purpose of this project is not neccesarily to create a high-performance solution. Instead, it aims to technically implement a more creative solution.
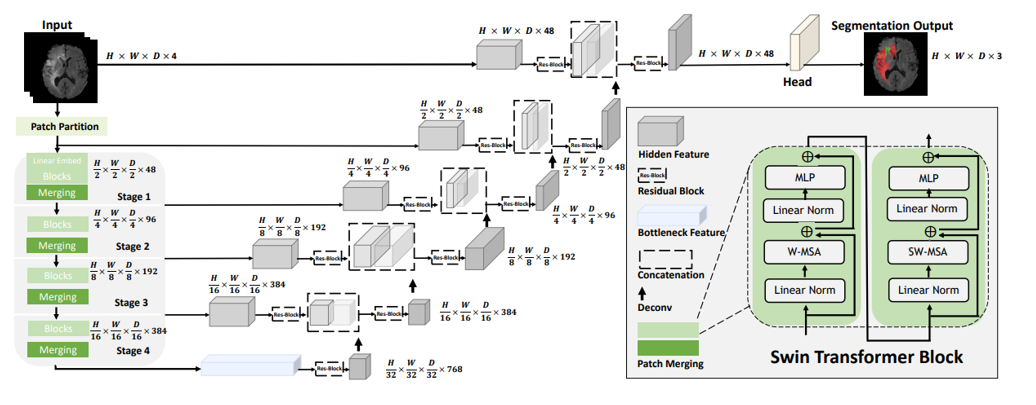
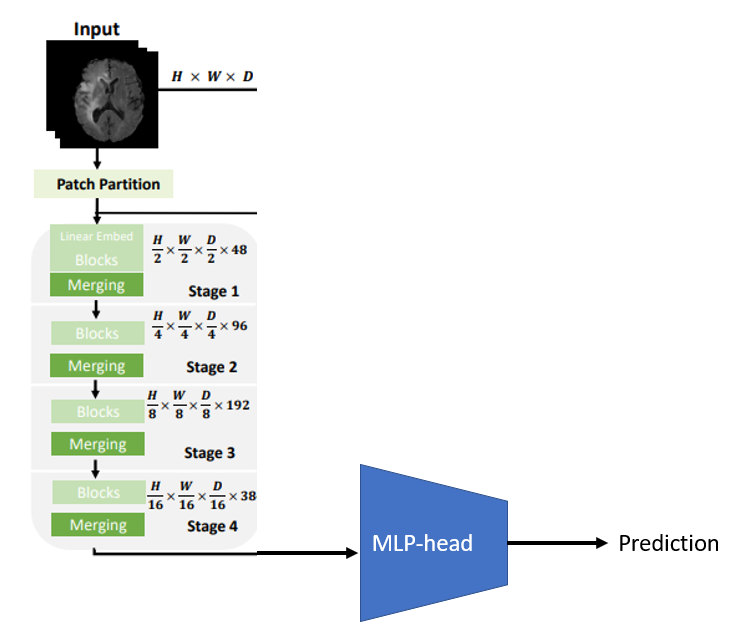
I used a pretrained Swin UNETR model, which was trained to segment brain tumours in CT scans by Hatamizadeh et al.. The architecture of this model is displayed in Figure 1. I used the pretrained ViT-encoder to extract features from the image and removed the rest of the model. Furthermore, I designed a MLP-head which was used to classify the clinical follow-up, based on these image features as shown in Figure 2.
Figure 1
Figure 2
The candidates dataset consisted of about 700,000 candidates, with approximately 1500 of those requiring follow-up. To speed up training times for this project, I decided to work with image patches with centered candidates. With regard to data storage, I selected all 1500 positive candidates and about 2000 negative candidates and saved the patches. The dataset was split 80:20 over a train and validation set. I coded data augmentations for all random possible flips and 90 degree rotations.
Initially, I saved the feature vectors which the Swin Encoder derived from the patches to efficiently train the MLP-head before training the entire model. This method did not produce any accurate results on the validation data. Consequently, I decided to train the Swin Encoder and MLP-head fully as a combined model.
After training for 30 epochs over the training data, the model reached an optimal classification accuracy of 85.4% over the validation set. Sensitivity was measured at 72.1% and specificity at 92.3% at a cutoff value of 0.5.
The train loss and validation accuracy are plotted in figure 3.
Figure 3
In this project, I designed and wrote the code for:
- The datasets which load the images patches or feature vectors and associated labels
- The scripts used to generate the image patches.
- The script used to save the feature vectors derived from these images
- The MLP-head
- The model combining the Swin Encoder and the MLP-head
- The training and validation loops
The only mostly external code was the code for the Swin Encoder. This derived from code for a Swin UNETR model, which I reduced to only include the ViT encoder.