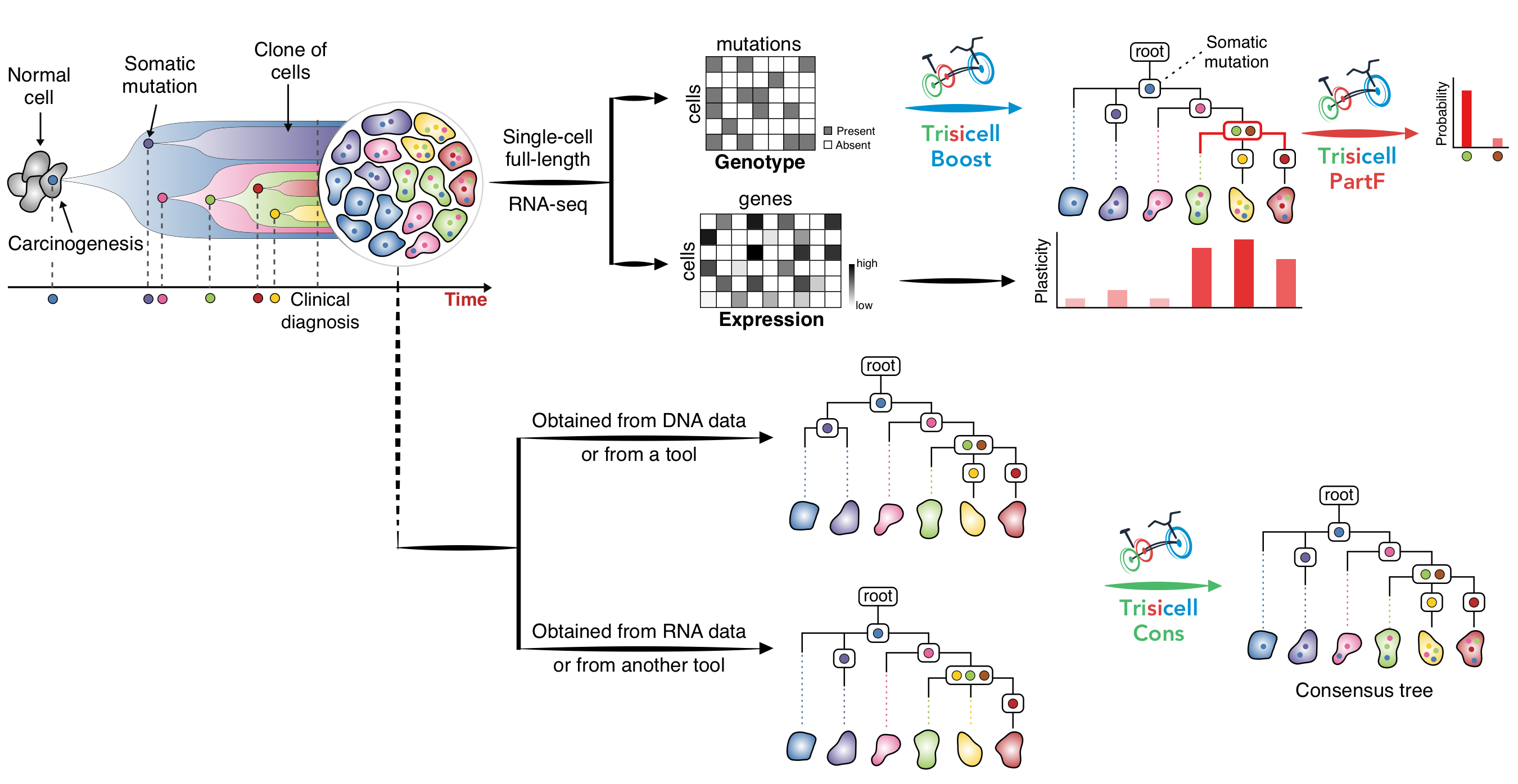
Trisicell (Triple-toolkit for single-cell intratumor heterogeneity inference), pronounced as "tricycle", is a new computational toolkit for scalable intratumor heterogeneity inference and evaluation from single-cell RNA, as well as single-cell genome or exome, sequencing data. Trisicell utilizes expressed SNVs and Indels to infer evolutionary relationships between genomic alterations and the cells that harbor them.
Feel free to submit an issue. Your help to improve Trisicell is highly appreciated.
Trisicell was developed in collaboration between the Cancer Data Science Laboratory (CDSL) and the Laboratory of Cancer Biology and Genetics (LCBG) at the National Cancer Institute (NCI).











