This is an umbrella repo describing statistical approximations of cell atlases.
The code is structured in separate repos:



Moreover, we are working on two additional repos:
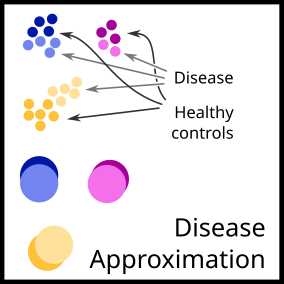
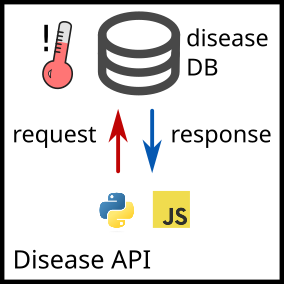
There are two main research directions:
- Cell atlases of healthy organisms across one billion years of evolution
- Single cell data from patients with diseases
The repos links for the healthy cell atlases are:
- Approximation (aka compression): https://github.com/fabilab/cell_atlas_approximations_compression
- API: https://github.com/fabilab/cell_atlas_approximations_API
- Natural Language Processing (NLP): https://github.com/fabilab/cell_atlas_approximations_NLP
- Human Interface: https://github.com/fabilab/cell_atlas_approximations_HI
- scquill (compress arbitrary data sets): https://github.com/fabilab/scquill
- SATURN-xfer (transfer learning): https://github.com/fabilab/SATURN-xfer (this repo is a fork of SATURN)
- Analysis (paper): https://github.com/fabilab/cell_atlas_approximations_analysis
The repos links for the disease cell atlases are:
- Disease approximation: https://github.com/fabilab/cell_atlas_approximations_disease_compression
- Disease API: https://github.com/fabilab/cell_atlas_approximations_disease_API