This repository contains the Python implementation of SEAM (Systematic Explanation of Attribution-based Mechanisms), an AI interpretation framework that systematically investigates how mutations reshape regulatory mechanisms. For an extended discussion of this approach and its applications, please refer to our manuscript, which we presented at the ICLR 2025 GEM Workshop:
- Seitz, E.E., McCandlish, D.M., Kinney, J.B., and Koo P.K. Decoding the Mechanistic Impact of Genetic Variation on Regulatory Sequences with Deep Learning. Workshop on Generative and Experimental Perspectives for Biomolecular Design, International Conference on Learning Representations, April 15, 2025. https://openreview.net/forum?id=PtjMeyHcTt
A bioRxiv preprint is also currently underway.
With Anaconda sourced, create a new environment via the command line:
conda create --name seam python==3.8*Next, activate this environment via conda activate seam, and install the following packages:
pip install seam-nnFinally, when you are done using the environment, always exit via conda deactivate.
If you have any issues installing SEAM, please see:
- https://seam-nn.readthedocs.io/en/latest/installation.html
- https://github.com/evanseitz/seam-nn/issues
For issues installing SQUID, the package used for sequence generation and inference, please see:
SEAM provides a unified interface for mechanistic interpretation of sequence-based deep learning models.
The framework takes as input a sequence-based oracle (e.g., a genomic DNN) and requires four key components to perform analysis:
-
Sequence Library (
numpy.ndarray): One-hot encoded sequences of shape (N, L, A), where:- N: Number of sequences
- L: Sequence length
- A: Number of features (e.g., 4 for DNA nucleotides)
-
Predictions/Measurements (
numpy.ndarray): Experimental or model-derived values of shape (N,1), corresponding to each sequence's functional output. -
Attribution Maps (
numpy.ndarray): Mechanistic importance scores of shape (N, L, A), quantifying the contribution of each position-feature pair to the sequence's function. These can be generated using various attribution methods: -
Clustering/Embedding (either):
- Hierarchical clustering linkage matrix (e.g., from
scipy.cluster.hierarchy.linkage) - Dimensionality reduction embedding of shape (N,Z), where Z is the number of dimensions in the embedded space
- Hierarchical clustering linkage matrix (e.g., from
These required files can be generated either externally or using SEAM's specialized modules (described below). Once provided, SEAM applies a meta-explanation approach to interpret the sequence-function-mechanism dataset, deciphering the determinants of mechanistic variation in regulatory sequences.
For detailed examples of how to generate these requirements using SEAM's modules and apply the analysis pipeline to reproduce key findings from our main manuscript, see the Examples section at the end of this document.
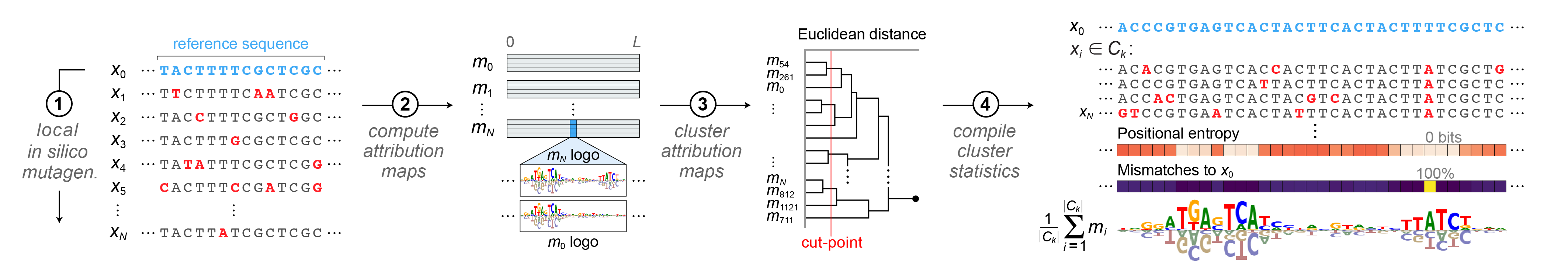
SEAM’s analysis pipeline is organized into modular components, with outputs from each stage feeding into the next. The Mutagenizer, Compiler, Attributer, and Clusterer modules generate core data products, which are integrated by the MetaExplainer to characterize each SEAM-derived mechanism. The Identifier module then builds on these outputs to annotate regulatory elements and quantify their combinatorial relationships.
-
Mutagenizer (from SQUID): Generates in silico sequence libraries through various mutagenesis strategies, including local, global, optimized, and complete libraries (supporting all combinatorial mutations up to a specified order). Features GPU-acceleration and batch processing for efficient sequence generation.
-
Compiler: Standardizes sequence analysis by converting one-hot encoded sequences to string format and computing associated metrics. Compiles sequences and functional properties into a DataFrame, with support for metrics such as Hamming distances and global importance analysis scores. Implements GPU-accelerated sequence conversion and vectorized operations.
-
Attributer: Computes attribution maps that quantify the base-wise contribution to regulatory activity. SEAM provides TensorFlow 2 GPU-accelerated implementations of Saliency Maps, IntGrad, SmoothGrad, and ISM. (For TF2, DeepSHAP is not yet optimized for GPU accelerated batch processing across the sequence library and requires external libraries
pip install kundajelab-shap==1andpip install deeplift). -
Clusterer: Computes mechanistic clusters and embeddings from attribution maps to identify distinct regulatory mechanisms. Supports hierarchical clustering (GPU-optimized), K-means, and DBSCAN algorithms, with optional dimensionality reduction (UMAP, t-SNE, PCA) for complementary interpretability.
-
MetaExplainer: The core SEAM module that integrates results to identify and interpret mechanistic patterns. Generates cluster-averaged attribution maps (shape: (L, A) for each cluster) and the Mechanism Summary Matrix (MSM), a DataFrame containing position-wise statistics (entropy, consensus matches, reference mismatches) for each cluster. Also implements background separation and provides visualization tools for sequence logos, attribution logos, and cluster statistics, with support for both PWM-based and enrichment-based analysis. Features GPU acceleration with CPU fallbacks.
-
Identifier: Analyzes cluster-averaged attribution maps in conjunction with the MSM to identify such properties as the precise locations of motifs and their epistatic interactions.
Google Colab examples for applying SEAM on previously-published deep learning models (e.g., DeepSTARR) and experimental datasets (e.g., PBMs) are available at the links below.
Note: Due to memory requirements for calculating distance matrices, Colab Pro may be required for examples using hierarchical clustering with their current settings.
- Local library to annotate all TFBSs and binding configurations
- DeepSTARR: Enhancer 20647 (Fig.2a)
- Local library with 30k sequences and 10% mutation rate | Integrated gradients; hierarchical clustering
- Expected run time: ~3 minutes on Colab A100 GPU
- Local library to reveal low-affinity motifs using background separation
- DeepSTARR: Enhancer 5353 (Fig.TBD)
- Local library with 60k sequences and 10% mutation rate | Integrated gradients; hierarchical clustering
- Expected run time: ~8.5 minutes on Colab A100 GPU
- Local library to explore mechanism space of an enhancer TFBS
- DeepSTARR: Enhancer 13748 (SFig.TBD)
- Local library with 100k sequence and 10% mutation rate | Saliency maps; UMAP with K-Means clustering
- Expected run time: ~3.9 minutes on Colab A100 GPU
- Combinatorial-complete library with empirical mutagenesis maps
- PBM: Zfp187 (Fig.TBD)
- Combinatorial-complete library with 65,536 sequences | ISM; Hierarchical clustering
- Expected run time: ~12 minutes on Colab A100 GPU
- Combinatorial-complete library with interactive mechanism space viewer
- PBM: Hnf4a (Fig.TBD)
- Combinatorial-complete library with 65,536 sequences | ISM; UMAP with K-Means clustering
- Expected run time: ~4.9 minutes on Colab A100 GPU
- Global library to compare mechanistic heterogeneity of an enhancer TFBS
- DeepSTARR: CREB/ATF (Fig.TBD)
- Global library with 100k sequences | Saliency maps: UMAP with K-Means clustering
- Expected run time: ~3.2 minutes on Colab A100 GPU
- Global library to compare mechanisms across different developmental programs
- DeepSTARR: DRE (Fig.TBD)
- Global library with 100k sequences | Saliency maps; UMAP with K-Means clustering
- Expected run time: ~2.7 minutes on Colab A100 GPU
- Global library to compare mechanisms associated with genomic and synthetic TFBSs
- DeepSTARR: AP-1 (Fig.TBD)
- Global library with 100k sequences | Integrated gradients; UMAP with K-Means clustering
- Expected run time: ~3.9 minutes on Colab A100 GPU
Python script examples are provided in the examples folder for locally running SEAM and exporting outputs to file. Some of these examples include models and/or attribution methods that are not compatible with the latest libraries supported by Google Colab, including:
- Local library to analyze foreground and background signals at human promotors and enhancers
- ChromBPNet: PPIF promoter/enhancer (Fig.3)
- Local library with 100k sequences and 10% mutation rate | {Saliency, IntGrad, SmoothGrad, ISM}; Hierarchical clustering
Additional dependencies for these Python examples may be required and outlined at the top of each script.
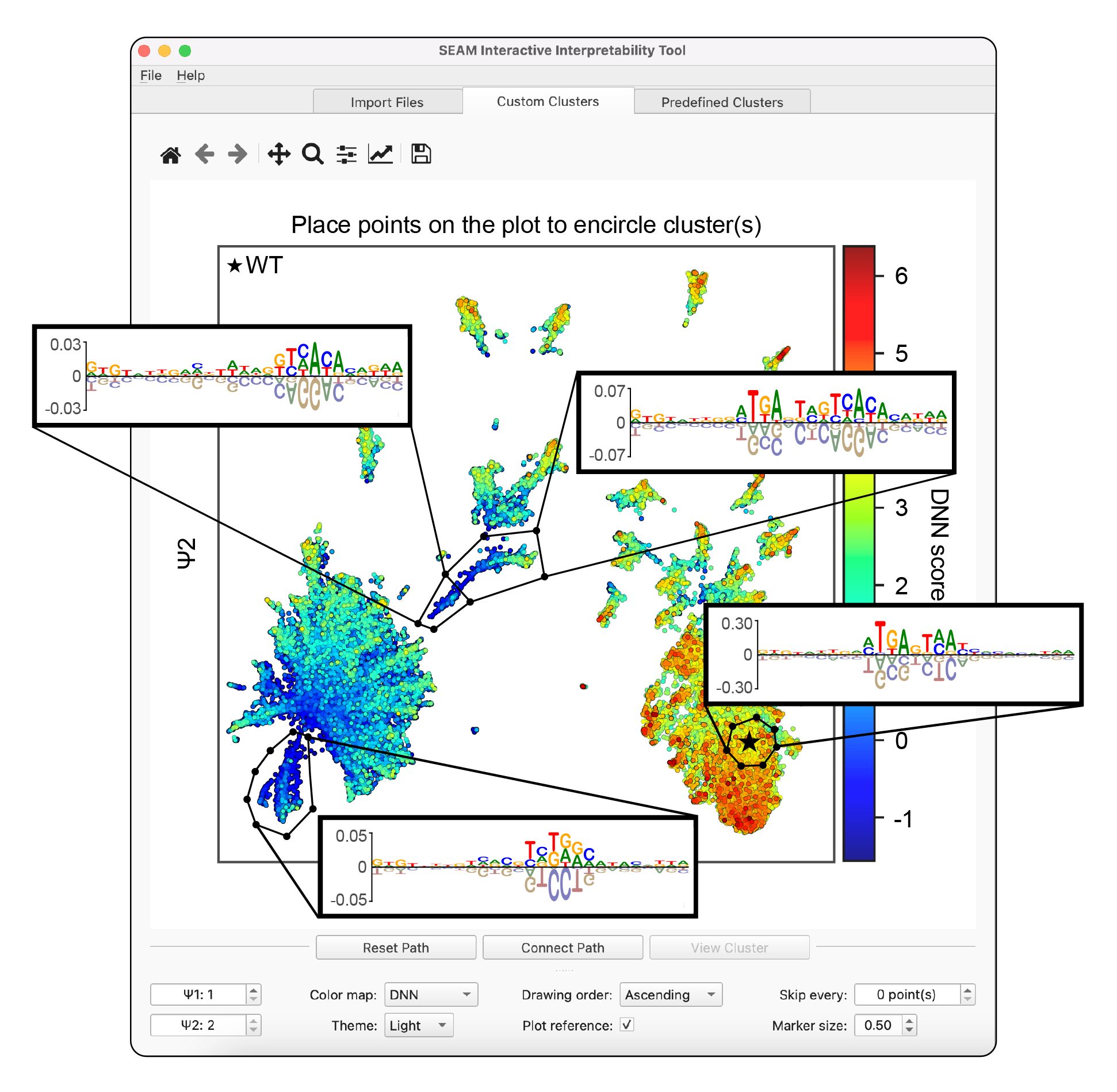
A graphic user interface (GUI) is available for dynamically interpretting SEAM results, allowing users to explore and analyze pre-computed inputs from the e. The GUI can be run using the command line interface from the seam folder via python seam_gui.py with the seam-gui environment activated (see below). The SEAM GUI requires pre-computed inputs that can be saved using the example scripts above. Instructions for downloading demo files for running the SEAM GUI are available in the seam/seam_gui_demo folder. A full walkthrough of the SEAM GUI using this demo dataset is available on YouTube.
SEAM GUI environment requires alternative imports to the default seam environment (above). The seam-gui environment can be installed following these steps:
conda create --name seam-gui python==3.8*Next, activate this environment via conda activate seam-gui, and install the following packages:
pip install --upgrade pip
pip install PyQt5
pip3 install --user psutil
pip install biopython
pip install scipy
pip install seaborn
pip install -U scikit-learn
pip install pysam
pip install seam-nn
pip install matplotlib==3.6To avoid conflicts, matplotlib==3.6 must be the last package installed
Finally, when you are done using the environment, always exit via conda deactivate.
If this code is useful in your work, please cite our paper.
bibtex TODO
Copyright (C) 2023–2025 Evan Seitz, David McCandlish, Justin Kinney, Peter Koo
The software, code samples and their documentation made available on this website could include technical or other mistakes, inaccuracies or typographical errors. We may make changes to the software or documentation made available on its web site at any time without prior notice. We assume no responsibility for errors or omissions in the software or documentation available from its web site. For further details, please see the LICENSE file.