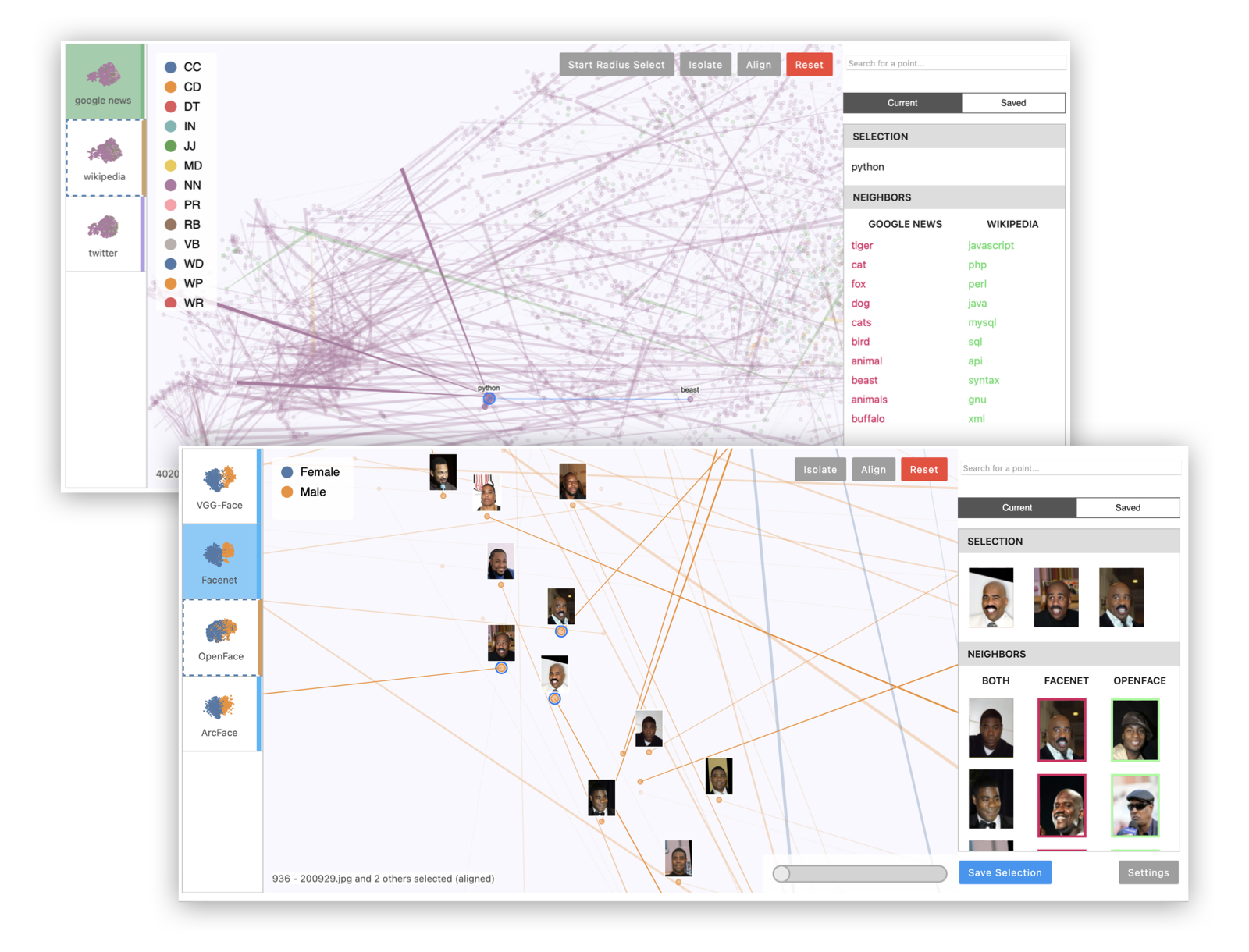
Emblaze is a Jupyter notebook widget for visually comparing embeddings using animated scatter plots. It bundles an easy-to-use Python API for performing dimensionality reduction on multiple sets of embedding data (including aligning the results for easier comparison), and a full-featured interactive platform for probing and comparing embeddings that runs within a Jupyter notebook cell. Read the documentation >
Compatibility Note: Emblaze has been tested using Python >= 3.7. If you are using JupyterLab, please make sure you are running version 3.0 or higher. NEW: The widget should work in VS Code and Colab environments thanks to using anywidget for platform-agnostic packaging. Please file a GitHub issue if you notice any compatibility problems.
Install Emblaze using pip:
pip install emblazeThe widget should work out of the box when you run jupyter lab (or open a notebook in VS Code or Colab) and follow the example code below.
Please see examples/example.ipynb to try using the Emblaze widget on the Boston housing prices or MNIST (TensorFlow import required) datasets.
Example 1: Multiple projections of the same embedding dataset. This can reveal areas of variation in the dimensionality reduction process, since tSNE and UMAP are randomized algorithms.
import emblaze
from emblaze.utils import Field, ProjectionTechnique
# X is an n x k array, Y is a length-n array
X, Y = ...
# Represent the high-dimensional embedding
emb = emblaze.Embedding({Field.POSITION: X, Field.COLOR: Y})
# Compute nearest neighbors in the high-D space (for display)
emb.compute_neighbors(metric='cosine')
# Generate UMAP 2D representations - you can pass UMAP parameters to project()
variants = emblaze.EmbeddingSet([
emb.project(method=ProjectionTechnique.UMAP) for _ in range(10)
])
# Compute neighbors again (to indicate that we want to compare projections)
variants.compute_neighbors(metric='euclidean')
w = emblaze.Viewer(embeddings=variants)
wExample 2: Multiple embeddings of the same data from different models. This is useful to see how different models embed data differently.
# Xs is a list of n x k arrays corresponding to different embedding spaces
Xs = ...
# Y is a length-n array of labels for color-coding
Y = ...
# List of strings representing the name of each embedding space (e.g.
# "Google News", "Wikipedia", "Twitter"). Omit to use generic names
embedding_names = [...]
# Make high-dimensional embedding objects
embeddings = emblaze.EmbeddingSet([
emblaze.Embedding({Field.POSITION: X, Field.COLOR: Y}, label=emb_name)
for X, emb_name in zip(Xs, embedding_names)
])
embeddings.compute_neighbors(metric='cosine')
# Make aligned UMAP
reduced = embeddings.project(method=ProjectionTechnique.ALIGNED_UMAP)
w = emblaze.Viewer(embeddings=reduced)
wExample 3: Visualizing image data with image thumbnails. The viewer will display image previews for each point as well as its nearest neighbors. (For text data, you can use TextThumbnails to show small pieces of text next to the points.)
# images is an n x 100 x 100 x 3 numpy array of 100x100 RGB images (values from 0-255)
images = ...
thumbnails = emblaze.ImageThumbnails(images)
w = emblaze.Viewer(embeddings=embeddings, thumbnails=thumbnails)
wYou can also visualize embeddings with multimodal labels (i.e. where some points have text labels and others have image labels) by initializing an emblaze.CombinedThumbnails instance with a list of other Thumbnails objects to combine.
See the documentation for more details on defining and configuring comparisons with Emblaze.
Clone repository, then install dependencies. (Note: you may find it easier to install SciPy using conda first - conda install scipy)
pip install -r requirements.txtInstall the python package.
pip install -e .In one terminal, cd into the client directory and then run vite. This will start a live reload service for the frontend. In another terminal, start a jupyter lab server and open a notebook to start the Emblaze viewer. When you edit the frontend code, you will need to reload the JupyterLab webpage to see the results. When you edit the backend code, you will need to restart the Jupyter Python kernel.
Install pdoc3: pip install pdoc3
Build documentation:
pdoc --html --force --output-dir docs --template-dir docs/templates emblazeBump the widget version in emblaze/_version.py, package.json, and pyproject.toml if applicable. Then build the notebook widgets:
vite build
Run the packaging script to generate the wheel for distribution:
pip install --upgrade build twine
python -m build
Upload to PyPI (replace <VERSION> with the version number):
twine upload dist/emblaze-<VERSION>*
- Svelte transitions don't seem to work well as they force an expensive re-layout operation. Avoid using them during interactions.