This repo contains the codebase for our paper:
DisProtEdit: Exploring Disentangled Representations for Multi-Attribute Protein Editing
📍 ICML 2025 Workshops (GenBio, FM4LS)
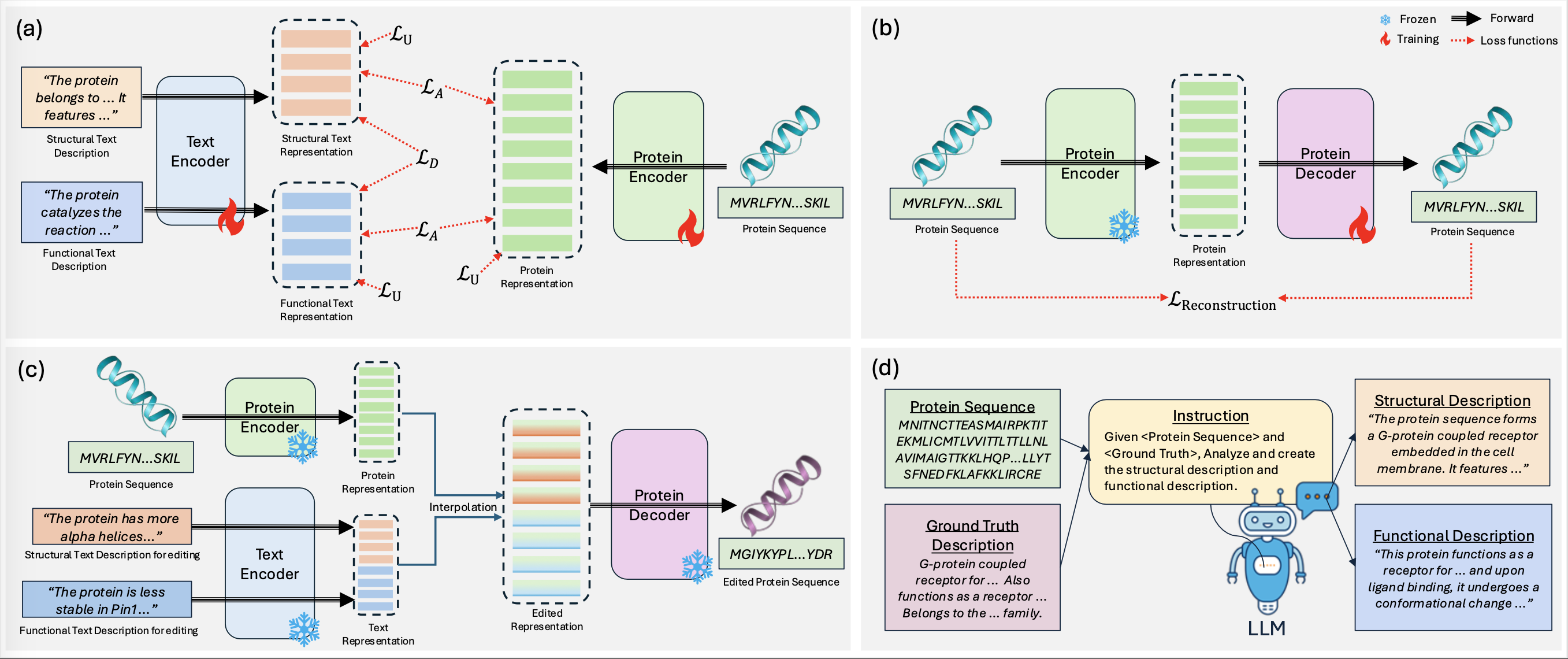
DisProtEdit is a protein editing framework that disentangles structural and functional properties using dual-channel natural language supervision. It learns modular latent representations aligned with protein sequences through a combination of alignment, uniformity, and angular MMD losses. Editing is performed via text modification, enabling interpretable and controllable edits to protein structure or function.
See https://tiger-ai-lab.github.io/DisProtEdit/ for more info.
- 2025 Jun 20: Released SwissProtDis Dataset, Editing benchmark, also the full training code.
- 2025 Jun 18: Paper available on Arxiv.
- 2025 Jun 17: Website created!
- 2025 Jun 11: DisProtEdit accepted to ICMLW GenBio.
- 2025 Jun 10: DisProtEdit accepted to ICMLW FM4LS.
We introduce SwissProtDis, a large-scale multimodal dataset containing:
- ~540,000 protein sequences
- Automatically decomposed structural and functional text descriptions from UniProt using GPT-4o
👉 https://huggingface.co/datasets/TIGER-Lab/SwissProtDis_500k
conda create -n disprot python=3.10
conda activate disprot
pip install -r requirements.txt
Training multimodal embeddings
./1_SFs.sh
./2_empty_SFs.sh
./2_sample_SFs.shAlternatively, you can run:
export OUTPUT_DIR="output/"
export PRETRAINED_DIR="output/SFs_AU_b24_gpt4o_500k_DisAngle10"
CUDA_VISIBLE_DEVICES=0,1,2,3\
python3 pretrain_step_01_SFs.py \
--protein_lr=1e-5 --protein_lr_scale=1 \
--text_lr=1e-5 --text_lr_scale=1 --CL_loss="EBM_NCE"\
--protein_backbone_model=ProtBERT_BFD --wandb_name="SFs_AU05_b24_gpt4o_500k"\
--epochs=10 --batch_size=24 --num_workers=0 --verbose \
--output_model_dir="$OUTPUT_DIR" --CL=0.0 --D=0.0 --U=0.5 --A=1.0 --dis_angle --ds_llm="gpt4o" --ds_name="500k"
python3 pretrain_step_02_empty_sequence_SFs.py \
--protein_backbone_model=ProtBERT_BFD \
--batch_size=16 --num_workers=4 \
--pretrained_folder="$PRETRAINED_DIR" \
--target_subfolder="pairwise_all"
python3 pretrain_step_02_pairwise_representation_SFs.py \
--protein_backbone_model=ProtBERT_BFD \
--batch_size=16 --num_workers=4 \
--pretrained_folder="$PRETRAINED_DIR" \
--target_subfolder="pairwise_all" \
--ds_llm="gpt4o"
Training decoder for editing task
./4_SFs.shAlternatively, you can run:
export PRETRAINED_DIR="output/SFs_AU_b24_gpt4o_500k_DisAngle10"
CUDA_VISIBLE_DEVICES=0\
python pretrain_step_04_decoder_SFs.py \
--batch_size=8 --lr=1e-4 --epochs=10 \
--decoder_distribution=T5Decoder \
--score_network_type=T5Base --wandb_name="SFs_AU_b24_gpt4o_500k_DisAngle10"\
--hidden_dim=16 --verbose \
--pretrained_folder="$PRETRAINED_DIR" \
--output_model_dir="$PRETRAINED_DIR"/step_04_T5 \
--target_subfolder="pairwise_all"Please see _datasets_and_checkpoints.
- The benchmark contains 196 protein inputs, suitable for protein editing on structure editing and functional editing.
- Please refer to
editing_dis_interpolation.py.
Multi-Attribute Protein Editing
./5_medit.shProtein Properties Prediction
./5_TAPE.shThe code is built upon TAPE in ProteinDT.
@misc{ku2025disproteditexploringdisentangledrepresentations,
title={DisProtEdit: Exploring Disentangled Representations for Multi-Attribute Protein Editing},
author={Max Ku and Sun Sun and Hongyu Guo and Wenhu Chen},
year={2025},
booktitle={ICML Workshop on Generative AI and Biology},
eprint={2506.14853},
archivePrefix={arXiv},
primaryClass={q-bio.QM},
url={https://arxiv.org/abs/2506.14853},
}This code is heavily built upon ProteinDT. we thank all the contributors for open-sourcing.