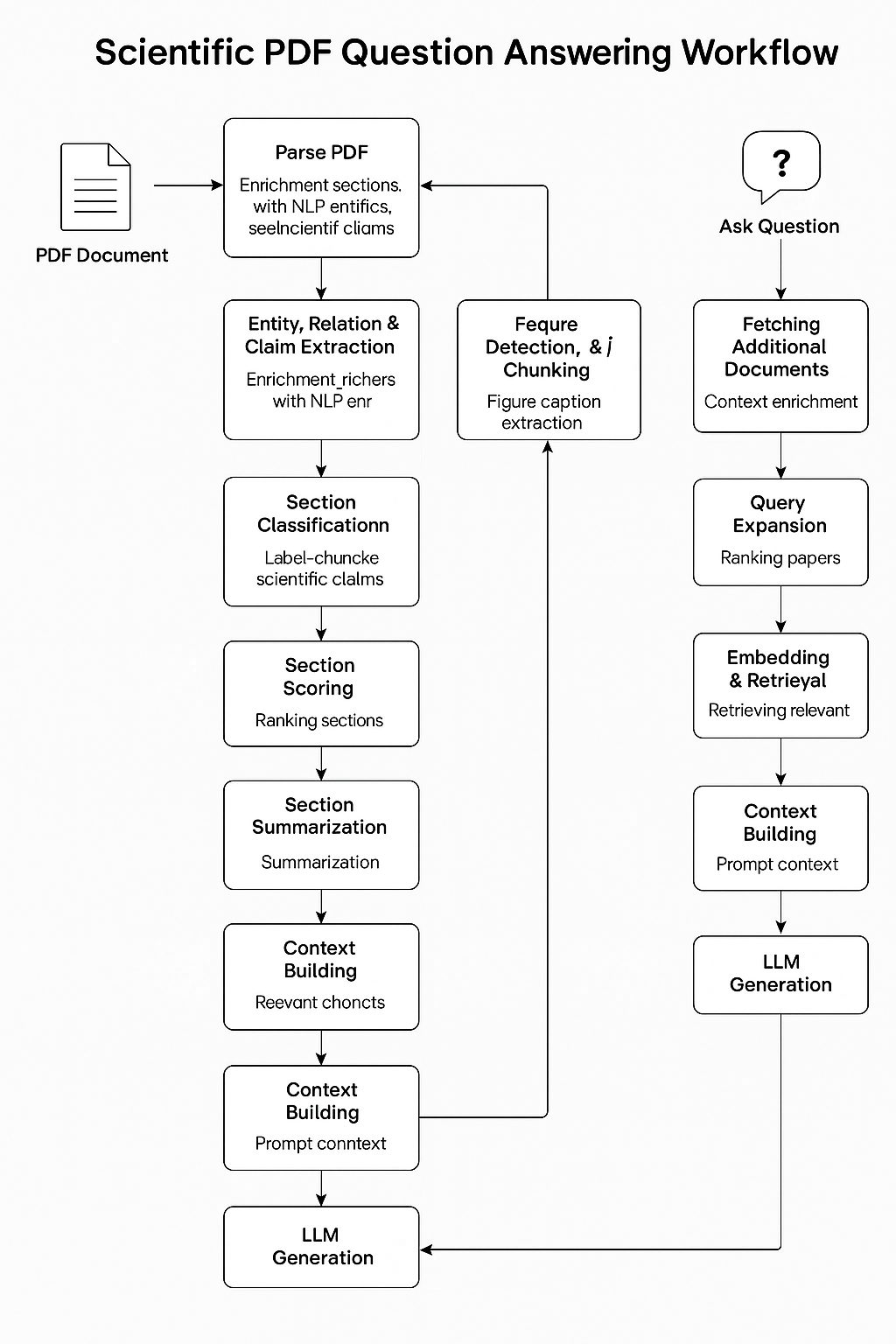
A modular and extensible Retrieval-Augmented Generation (RAG) architecture designed for automated scientific document comprehension and domain-specific question answering. The pipeline integrates hierarchical PDF decomposition, fine-grained entity and relation extraction, citation graph analytics, semantically-aware vector retrieval via hybrid embedding strategies, and local LLM-driven response synthesis, enabling end-to-end reasoning over scholarly corpora.
- Scientific PDF parsing with section/figure/reference extraction
- Named entity, relation & claim detection using spaCy & SciSpacy
- Section classification via fine-tuned BERT
- Layout-aware figure detection using LayoutLM
- Semantic query expansion with BERT
- Citation graph construction and scoring
- Embedding & hybrid retrieval with ChromaDB
- Prompt construction and budgeted context building
- Final LLM answer generation via local Mistral (GGUF format)
- Achieved F1 score of 0.9954 while transfer learning the Reference Parsing model.
- Achieved F1 score of 0.7994 while transfer learning the Section Classification model.
- Function:
parse_pdf(pdf_to_document.py) - Uses Grobid for metadata, tokenizes content, and prepares chunks
- Entity Extractor:
ScientificEntityExtractor - Relation Extractor:
PatternRelationExtractor - Claim Extractor:
ScientificClaimDetector - Adds entities, relations, and claims to section metadata
- Function:
classify_sections - Label sections (Introduction, Methods...)
- Chunk and embed text for ChromaDB
- Functions:
detect_figures_in_pdf,convert_figures_to_chunks - Use LayoutLM to detect captions, convert to retrievable chunks
- In
pipeline_runner.py: normalizes raw references to title, authors, journal, etc.
- Function:
score_sections - Based on citation count, recency, and word count
- Function:
summarize_sections - One-line summaries per section
- Function:
fetch_additional_documents - Stubbed integration with arXiv / Semantic Scholar
- Class:
BERTQueryExpander - BERT-based semantic keyword expansion
- Class:
CitationNetwork - Builds citation graph and extracts influence clusters
- Class:
FieldSpecificSearchOptimizer - Ranks related papers for context enrichment
- Classes:
ScientificEmbedding,Retriever - Retrieve top-k relevant chunks from ChromaDB using a hybrid search
- Class:
ContextBuilder - Assembles context string for LLM with figure-aware logic
- Functions:
build_safe_prompt,safe_generate,LLMRunner - Prepares the prompt and runs local LLM (Mistral) to get answers
- In:
pipeline_runner.py - Answer sent to
stdoutas:LLM Answer: ...
| Step | Function/Class | Input | Output | Role |
|---|---|---|---|---|
| 1 | parse_pdf | PDF path | Document dict | PDF → structured text/metadata |
| 1.5 | Entity/Relation/Claim Extractors | Section text | Entities, relations, claims | NLP enrichment |
| 2 | classify_sections | Sections | Labeled + embedded sections | Section classification |
| 2.5 | detect_figures_in_pdf | PDF + tokenizer | Figure chunks | Figure caption detection |
| 2.6 | pipeline_runner.py | Raw references | Cleaned references | Metadata normalization |
| 3 | score_sections | Sections | Scored list | Rank importance |
| 4 | summarize_sections | Sections | One-line summaries | Context hinting |
| 5 | fetch_additional_documents | Paper title | Related papers | Context expansion |
| 5.1 | BERTQueryExpander | Keywords | Similar terms | Improve recall |
| 5.2 | CitationNetwork | Papers | Graph & communities | Citation analysis |
| 5.3 | FieldSpecificSearchOptimizer | Titles | Relevance scores | Re-ranking |
| 6 | Retriever + Embeddings | Query | Top-k chunks | Semantic retrieval |
| 7 | ContextBuilder | Chunks + Query | Context string | Prompt assembly |
| 8 | LLMRunner | Prompt + context | Final answer | LLM inference |
| 9 | pipeline_runner.py | LLM answer | stdout | UI output |
demo.mp4
- Launch the Streamlit app via
streamlit run app.py - Paste the path to the PDF , ask a question, see the result
# Clone the repo
git clone https://github.com/<your-org>/ResearchRAG-End-to-End-Scientific-Research-Assistant-using-RAG
# Navigate into the project
cd ResearchRAG-End-to-End-Scientific-Research-Assistant-using-RAG
# Create virtual environment
python -m venv LOLvenv && source LOLvenv/bin/activate # for Linux/macOS
# OR for Windows:
python -m venv LOLvenv && LOLvenv\Scripts\activate
# Install dependencies
pip install -r requirements.txt
# Download LLM model (Mistral 7B GGUF)
# Save it inside: models/llm_gguf/
# File: mistral-7b-instruct-v0.1.Q4_K_M.gguf
# Run GROBID container (required for metadata extraction)
docker run -t --rm -p 8070:8070 lfoppiano/grobid:latest
# Run the main app
python app.pyWe fine-tuned a LayoutLMv1 model on the PubLayNet dataset to detect figure captions and their bounding boxes in scientific PDFs, enabling layout-aware chunking of figure content.
A BERT base model was fine-tuned on a 5K subset of S2ORC for scientific section classification (Introduction, Methods, Results, etc.). This significantly improved accuracy over rule-based or zero-shot methods.
- CORD-19: Used for testing end-to-end scientific PDF parsing and question answering.
- S2ORC (5K subset): Used to fine-tune our section classification model and evaluate semantic embedding quality.
- PubLayNet: Utilized for training our LayoutLM model to achieve accurate figure detection in scientific PDFs.
- arXiv Papers: Sampled for testing document expansion, similarity ranking, and citation graph analysis.
| Task | F1 Score |
|---|---|
| Section Classification | 79.94% |
| Reference Parsing | 99.54% |
- Grobid: for metadata parsing
- Hugging Face: for pretrained scientific models
- ChromaDB: for open-source vector DB
- Mistral: local LLM inference (GGUF)