The dataset is from Kaggle called Fetal Health Classification, which was cleaned and visualized.
This dataset is provided with the aim to prevent child and maternal mortality.
Quote from the dataset page:
Reduction of child mortality is reflected in several of the United Nations' Sustainable Development Goals and is a key indicator of human progress. The UN expects that by 2030, countries end preventable deaths of newborns and children under 5 years of age, with all countries aiming to reduce under‑5 mortality to at least as > low as 25 per 1,000 live births.
Parallel to notion of child mortality is of course maternal mortality, which accounts for 295 000 deaths during and following pregnancy and childbirth (as of 2017). The vast majority of these deaths (94%) occurred in low-resource settings, and most could have been prevented.
In light of what was mentioned above, Cardiotocograms (CTGs) are a simple and cost accessible option to assess fetal health, allowing healthcare professionals to take action in
order to prevent child and maternal mortality. The equipment itself works by sending ultrasound pulses and reading its response, thus shedding light on fetal heart rate (FHR), fetal movements, uterine contractions and more.
This dataset contains 2126 records of 22 features extracted from Cardiotocogram exams, which were then classified by three expert obstetritians into 3 classes:
Normal(represented by target variable 1)Suspect(represented by target variable 2)Pathological(represented by target variable 3)
| No | Feature | Description |
|---|---|---|
| 1 | baseline_value |
Baseline Fetal Heart Rate (FHR) |
| 2 | accelerations |
Number of accelerations per second |
| 3 | fetal_movement |
Number of fetal movements per second |
| 4 | uterine_contractions |
Number of uterine contractions per second |
| 5 | light_decelerations |
Number of LDs per second |
| 6 | severe_decelerations |
Number of SDs per second |
| 7 | prolongued_decelerations |
Number of PDs per second |
| 8 | abnormal_short_term_variability |
Percentage of time with abnormal short term variability |
| 9 | mean_value_of_short_term_variability |
Mean value of short term variability |
| 10 | percentage_of_time_with_abnormal_long_term_variability |
Percentage of time with abnormal long term variability |
| 11 | mean_value_of_long_term_variability |
Mean value of long term variability |
| 12 | histogram_width |
Width of the histogram made using all values from a record |
| 13 | histogram_min |
Histogram minimum value |
| 14 | histogram_max |
Histogram maximum value |
| 15 | histogram_number_of_peaks |
Number of peaks in the exam histogram |
| 16 | histogram_number_of_zeroes |
Number of zeroes in the exam histogram |
| 17 | histogram_mode |
Hist mode |
| 18 | histogram_mean |
Hist mean |
| 19 | histogram_median |
Hist Median |
| 20 | histogram_variance |
Hist variance |
| 21 | histogram_tendency |
Histogram trend |
| 22 | fetal_health |
Fetal health: 1 - Normal ; 2 - Suspect ; 3 - Pathological |
Important
Only the notebook titled 0. Transforming data.ipynb is ran.
It standizes the target variable to make it more easier on the model.
Other notebooks are provided as an illustration of what was done and the conclusions that led to the final model.
The data was first loaded into a Jupyter notebook 1. Data preparation and data cleaning.
Upon inspection it was concluded that no cleaning was necessary as the data was very clean and organized, nor was there any need of standardizing column names and values, yet the code was included as a part of a streamlined solution that can be applied to other projects. In this notbook the most important step was temporarily converting a target variable from float to string
Next step was running the notebook 2. EDA, feature importance analysis.
In this notebooks certain visualizations were made to get a feel for the data to be worked with as well as feature importance analysis in form of viewing the correlations of features with each other as well as seing if any particular feature stood out. Techniques like mutual information, risk rate and correlation matrix were used.
Finally a notebook 3. Model selection process is ran to test the dummy models before they are converted to a script in form of a train.py.
The script train.py incorporates the tested model and creates a model from tabular data (no need for label encoding or dict vectorization, as all features are numeric) using the Decision Tree module from scikit-learn, a non-parametric supervised learning algorithm traditionally used for regression and classification tasks.
In this case the aim of the model was to predict the target variable of three classes using the multi-classification method. The model is then pickled for later use.
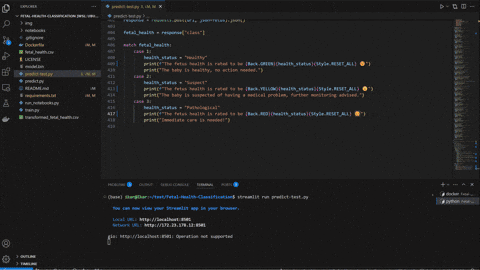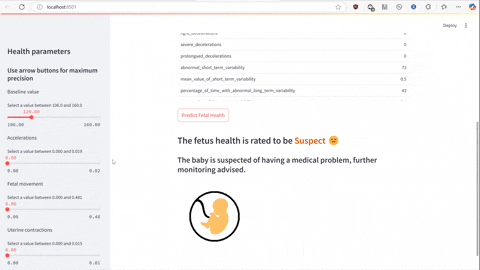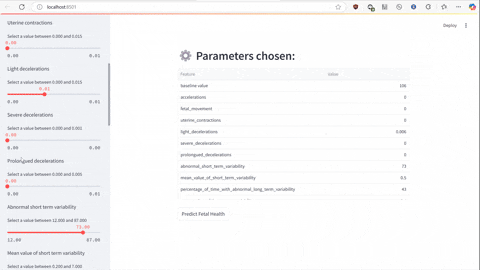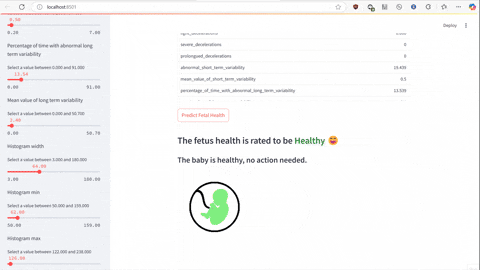
The script predict.py uses Flask and gunicorn to serve the unpickled model on local host network to which predict-test.py sends a POST request using JSON serialized data to make the prediction of a class the data falls under.
Important!
This project was developed on WSL (Windows subsystem for Linux).
It is recommended if on Windows to install it using a guide or running it on UNIX/Linux environment.
You will also need Docker.
- cd into your desired folder and download the project
git clone https://github.com/MortalWombat-repo/Fetal-Health-Classification.git
- cd into the folder
cd Fetal-Health-Classification
- build docker container
docker build -t fetal_health .
- run the container with the exposed port (very important, don't forget the p flag)
docker run -p 9696:9696 fetal_health
- open a new terminal tab and send a post request
python predict-test.py
- send a post request from a streamlit frontend
streamlit run predict-test.py
-
when done return to the window with the server and CTRL/Command + C to stop the server
-
exit the directory and delete the project
cd ..
rm -rf Fetal-Health-Classification
For viewing fullscreen click on the link.
Video Link
For viewing fullscreen click on the link.
Video Link
Source authors:
Ayres-de-Campos, D., Bernardes, J., Garrido, A., Marques-de-Sá, J. and Pereira-Leite, L. (2000), Sisporto 2.0: A program for automated analysis of cardiotocograms. J. Matern. Fetal Med., 9: 311-318. https://doi.org/10.1002/1520-6661(200009/10)9:5<311::AID-MFM12>3.0.CO;2-9
Link
Kaggle dataset author:
andrewmvd (Larxel)
Link