-
|
@a-ignatieva, who has started to investigate using tree sequences to look at SARS-CoV2, pointed out a useful thing that she (and others) might want to do, which is to be able to "collapse" all the descendants of an MRCA as long as the genomes descendant from that MRCA are all clonal (i.e. have undergone no detectable recombination). This would be quite easy using An alternative requirement might be to identify ancestral nodes for which all the descendant samples are the same (i.e. there might have been recombination between the descendant lineages but not outside, defining what we might call a "monophyletic" clade in a tree sequence). I wonder if there's a nice way to identify all these nodes? |
Beta Was this translation helpful? Give feedback.
Replies: 7 comments 20 replies
-
|
Good question! I wonder is there some incremental algorithm where we work from left to right? I.e, all nodes start as clonal and then as we work through the edge_diffs we mark nodes as being non clonal if they are a parent in an edge diff, or ancestral to a parent? Must be something like that, right? |
Beta Was this translation helpful? Give feedback.
-
|
We just need to ask which nodes have only one parent in the edge table, right? So, something like: I happen to have a utility for that in pyslim: |
Beta Was this translation helpful? Give feedback.
-
|
Is this the sort of thing we eventually want as a function in tskit, or is a (tested?) recipe here sufficient in this case? |
Beta Was this translation helpful? Give feedback.
-
|
I think this does the job: import numpy as np
import msprime
ts = msprime.sim_ancestry(5,
recombination_rate=0.002, sequence_length=100, random_seed=53)
print("ORGINAL TS")
print(ts.draw_text())
tables = ts.dump_tables()
index = np.logical_and(
tables.edges.left == 0, tables.edges.right == tables.sequence_length)
tables.edges.set_columns(
left=tables.edges.left[index],
right=tables.edges.right[index],
parent=tables.edges.parent[index],
child=tables.edges.child[index])
print("ONLY FULL SPAN EDGES")
ts2 = tables.tree_sequence()
print(ts2.draw_text())
print("SIMPLIFIED (but different IDs)")
ts3, node_map = ts2.simplify(map_nodes=True)
reverse_map = {mapped: j for j, mapped in enumerate(node_map) if mapped != -1}
print(ts3.draw_text())
tree = ts3.first()
mrcas = [root for root in tree.roots]
original_nodes = [reverse_map[root] for root in mrcas]
print("TRIMMED")
ts_trimmed = ts.simplify(original_nodes)
print(ts_trimmed.draw_text())Giving: Right? Surely removing all the non-full span edges is what we want, by definition. The rest is just fiddling to get rid of some annoying topology and mapping of node IDs. |
Beta Was this translation helpful? Give feedback.
-
|
I think the following is the right logic for the left-to-right algorithm that @jeromekelleher suggested. Probably worth checking against @petrelharp 's suggestion? def clonal_mrcas(ts):
# full length nodes without recombination will not be a parent in the edges in or out, apart from at the start
for interval, edges_out, edges_in in ts.edge_diffs():
if interval.left==0:
is_full_length_clonal = np.ones(ts.num_nodes, dtype=bool) # all nodes start as clonal
else:
for e in edges_in:
is_full_length_clonal[e.parent] = False
for e in edges_out:
is_full_length_clonal[e.parent] = False
clonal_nodes = np.where(is_full_length_clonal)[0]
tables = ts.dump_tables()
edges = tables.edges
# only keep edges where both the child and the parent are full-length clonal nodes
keep_edge = np.logical_and(np.isin(edges.child, clonal_nodes), np.isin(edges.parent, clonal_nodes))
tables.edges.set_columns(
left = tables.edges.left[keep_edge],
right=tables.edges.right[keep_edge],
parent=tables.edges.parent[keep_edge],
child=tables.edges.child[keep_edge],
)
ts = tables.tree_sequence()
print("Debug: show the clonal subtrees", ts.draw_text(), sep="\n")
assert ts.num_trees == 1
return ts.first().rootsIt works OK on my previous example. Here's another: ts = msprime.simulate(8, recombination_rate=1, random_seed=321)
print("Original ts", ts.draw_text(), sep="\n")
clonal_nodes = clonal_mrcas(ts)
print(f"Subtrees under nodes {clonal_nodes} are clonal")
reduced_ts, node_map = ts.simplify(clonal_nodes, map_nodes=True)
print(
"The tree seq with clonal subtrees removed",
"(drawn using the original labels)",
reduced_ts.draw_text(node_labels = {n:f"{i}" for i, n in enumerate(node_map)}),
sep="\n",
)Giving |
Beta Was this translation helpful? Give feedback.
-
|
Gah, I'm not sure the selected answer here does actually work. Here's a counterexample: tables = tskit.Tree.generate_comb(5).tree_sequence.dump_tables()
e = np.where(tables.edges.child == 2)[0][0]
tables.edges[e] = tables.edges[e].replace(right=0.5)
tables.sort()
ts = tables.tree_sequence()
print(ts.draw_text())If we run the code above, we erroneously identify the root (8) as the top of a clonal subtree: clonal_nodes = clonal_mrcas(ts)
print(f"Subtrees under nodes {clonal_nodes} are clonal")
reduced_ts, node_map = ts.simplify(clonal_nodes, map_nodes=True)
print(
"The tree seq with clonal subtrees removed",
"(drawn using the original labels)",
reduced_ts.first().draw_text(node_labels = {n:f"{i}" for i, n in enumerate(node_map)}),
sep="\n",
)The only clonal subtree here, IMO, is below node 5 (although we should also be reporting nodes 0, 1, and 2 as clonal) |
Beta Was this translation helpful? Give feedback.
-
|
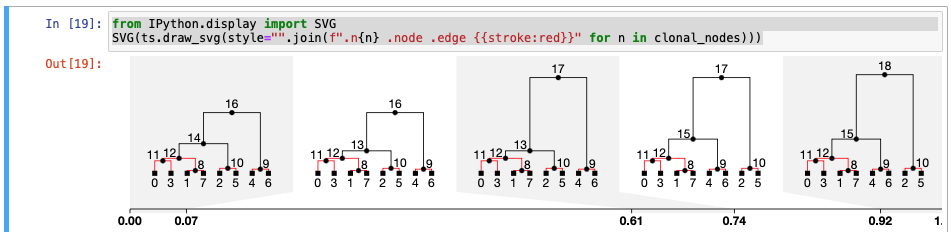
An updated answer based on my previous comments. It's more complicated because we need to follow up the trail from existing non-clonal edges and mark all their ancestors as non-clonal too. import numpy as np
def clonal_mrcas(ts, most_recent=True):
"""
Identify the nodes in a tree sequence which define clonal subtrees (i.e. in which the
samples descending from that node are identical and show identical relationships
to each other over the entire tree sequence). This includes, at its limit, nodes
with only a single descendant sample.
:param bool most_recent: If True, and the clonal node is a unary node, return IDs of the
most recent node that defines the clonal subtree. In this case, the returned IDs represent
cases where the node is either a tip or a coalescent point.
:return: a list of nodes defining subtrees which are constant over the entire tree sequence
:rtype: list
"""
for interval, edges_out, edges_in in ts.edge_diffs():
if interval.left==0:
is_full_length_clonal = np.ones(ts.num_nodes, dtype=bool) # all nodes start as clonal
else:
for e in edges_in:
is_full_length_clonal[e.parent] = False
for e in edges_out:
is_full_length_clonal[e.parent] = False
clonal_nodes = np.where(is_full_length_clonal)[0]
tables = ts.dump_tables()
edges = tables.edges
# only keep edges where both the child and the parent are full-length clonal nodes
keep_edge = np.logical_and(np.isin(edges.child, clonal_nodes), np.isin(edges.parent, clonal_nodes))
tables.edges.set_columns(
left = tables.edges.left[keep_edge],
right=tables.edges.right[keep_edge],
parent=tables.edges.parent[keep_edge],
child=tables.edges.child[keep_edge],
)
clonal_ts = tables.tree_sequence()
assert clonal_ts.num_trees == 1
# Also remove all the edges ascending from removed edges
tree = clonal_ts.first()
non_clonal_ancestors = set()
deleted_edges = np.logical_not(keep_edge)
for u in np.unique(ts.edges_parent[deleted_edges]):
while u != tskit.NULL and u not in non_clonal_ancestors:
non_clonal_ancestors.add(u)
u = tree.parent(u)
non_clonal_ancestors = np.array(list(non_clonal_ancestors))
tables = clonal_ts.dump_tables()
remove_edge = np.isin(tables.edges.parent, non_clonal_ancestors)
tables.edges.replace_with(tables.edges[np.logical_not(remove_edge)])
clonal_ts = tables.tree_sequence()
tree = ts.first(sample_lists=True)
clonal_tree = clonal_ts.first(sample_lists=True)
clonal_nodes = []
for root in clonal_tree.roots:
# Clonal trees should have the same set of samples underneath as in the original tree
assert set(tree.samples(root)) == set(clonal_tree.samples(root))
u = root
if most_recent:
# decend to the first coalescent node (i.e. MRCA)
while clonal_tree.num_children(u) == 1:
u = clonal_tree.children(u)[0]
clonal_nodes.append(u)
return clonal_nodes |
Beta Was this translation helpful? Give feedback.
An updated answer based on my previous comments. It's more complicated because we need to follow up the trail from existing non-clonal edges and mark all their ancestors as non-clonal too.