Integrating datasets with multiple biological replicates and multiple conditions? #5702
Replies: 2 comments 3 replies
-
Have you found a solution to this? I'm thinking of doing the same thing. Could you cite the paper? |
Beta Was this translation helpful? Give feedback.
-
|
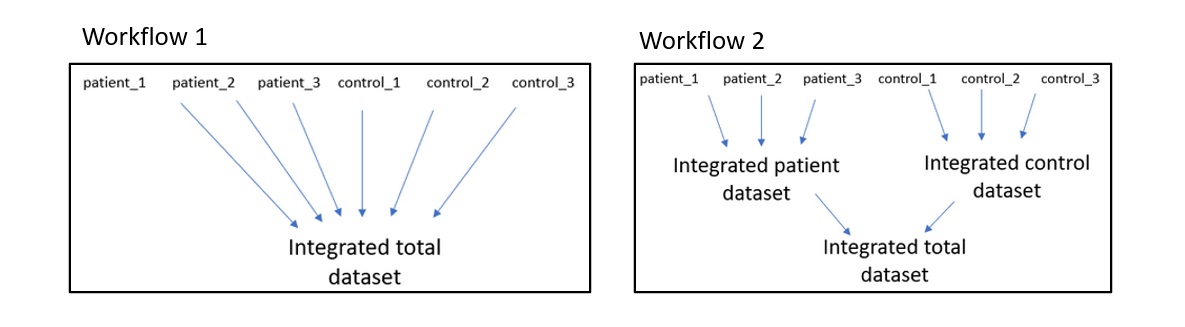
Not a dev. Workflow 2 is reasonable. This is because the underlying assumption is that the drug perturbations will be common to all the Treated samples. Therefore it is reasonable to first integrate controls and then the treatments separately, generating 2x Seurat objects and then integrating the two together. You don't need to worry about it tho Seurat's got your back, it automates this process for you. Essentially this can also be done using: You can then treat data.integrated as an integrated dataset where certain data are used as reference samples in this case samples 1, 3, 5, 7 and 9. Enjoy! 🐲 |
Beta Was this translation helpful? Give feedback.
Uh oh!
There was an error while loading. Please reload this page.
-
Hello,
I am currently analyzing a public single cell dataset.
It contains datasets for multiple biological replicates across 2 conditions: patients and controls.
To integrate the data I took all samples and integrated them in 1 go using the FindIntegrationAnchors() and IntegrateData() functions from Seurat as shown in "workflow 1" in the image below.
However when my resulting UMAP's and clusters did not look alot like the figures shown in the paper I looked at their code and found out they did the integration in 2 steps as shown in "workflow 2" in the image above.
I have never come across someone doing it like this. But I am quite new to analyzing single cell datasets so now I am wondering:
Is it "correct" to do the integration in 2 steps or might this have adverse effects?
Looking forward to hear your thoughts and thanks in advance!
Thomas
Beta Was this translation helpful? Give feedback.
All reactions