-
|
Hello. I'm new to scRNA and I've been looking at the guided PBMC tutorial and using that as a guide on doing analysis for data I've generated from 10x. I had a question regarding the part of the guide where the data is filtered based on QC metrics: pbmc <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5) If I try to apply "nFeature_RNA < 2500", that eliminates almost all of my cells. I was wondering if it's normal for 10x data to have such high nFeature values? |
Beta Was this translation helpful? Give feedback.
Replies: 3 comments 1 reply
-
|
Do you have a distribution plot for all the 3 features that you can share? |
Beta Was this translation helpful? Give feedback.
-
|
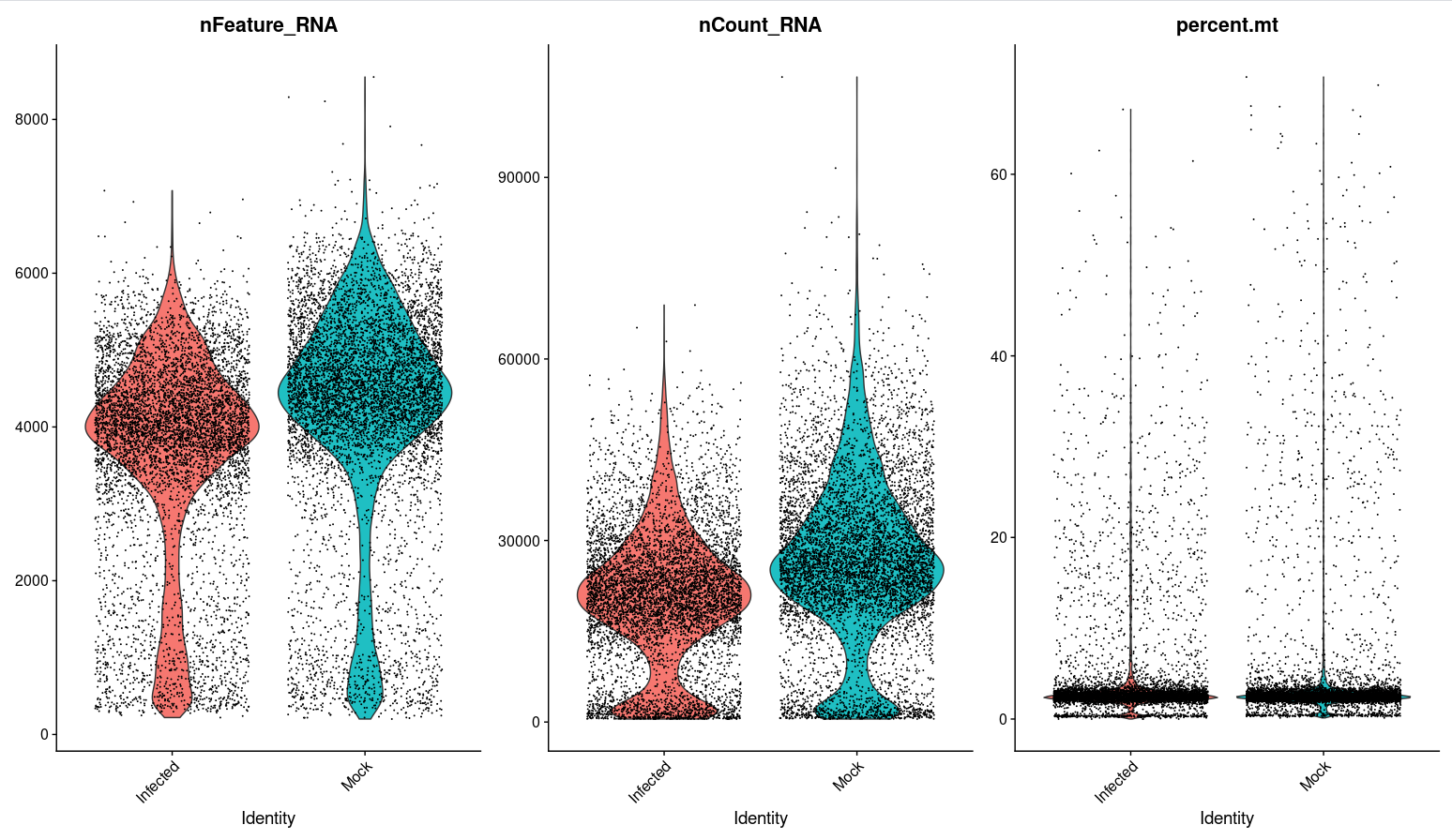
Here's my violin plot for the 3 features. |
Beta Was this translation helpful? Give feedback.
-
|
Got it. Thanks! |
Beta Was this translation helpful? Give feedback.
Got it. Thanks!