Replies: 1 comment
-
|
Okay, I found out what is the problem, nothing to do with the tiling. It turns out the output from file339 onwards has an additional six scalar constants in each netcdf file. I am now doing the following: That seems to be sufficient. I did not even have to drop those six constants from file339 onwards. Just splitting the datasets and concatenating later works. [Probably related to this issue?] |
Beta Was this translation helpful? Give feedback.
0 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
I am facing a weird error where depending on how I subset the same number of files, I end up getting NaN values in one case and not the other. Unfortunately, I don't know how to create a MVE for this issue. Here is what I am trying to do:
I have a set of netcdf files one per day for the year 2014 (365 total), say, files = {file0, file1,....file364}. The file has a bunch of variables with dimensions
(time,latitude,longitude). Thetimedimension is adatetime64object.I create a couple of datasets splitting the 365 files into two sets: fileA={file0...,file338} and fileB={file339....file364}.
When I plot the timeseries of a variable at some
(lat,lon), I get something like the picture below. The x-axis in the left plot starts from Jan. 1, as it should. The plot on the right shows continuity with the left plot, which also makes sense.[The titles for the plots should read "first 339 files" and "files 340-365".]
Here is what the timeseries for the same variable at the same (lat,lon) looks like:
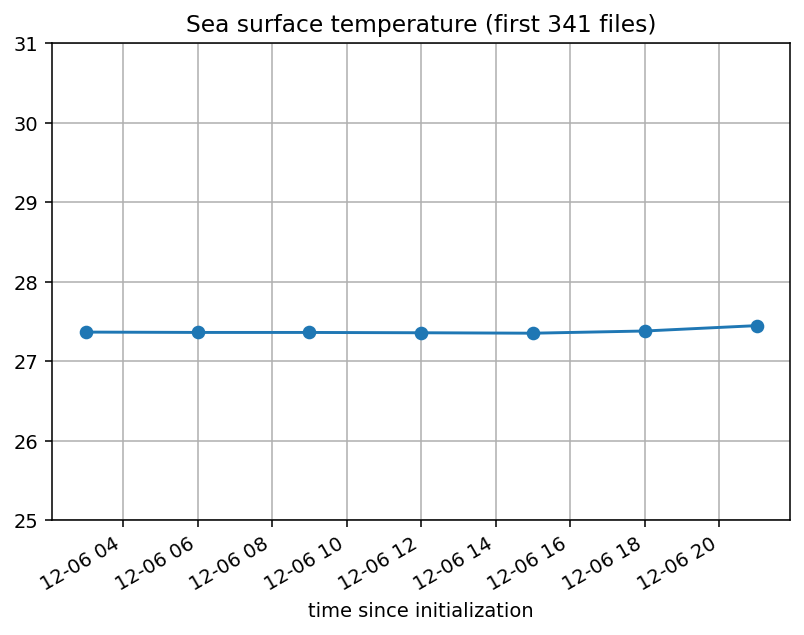
The x-axis now has shrunk remarkably and incorrectly starts from Dec. 6th, not Jan. 1st. This is because all values prior to Dec. 6th have been (incorrectly) read in as
NaN! But the first plot shows values prior to Dec. 6th are notNaN.I am baffled why my results should pivot on the inclusion of one extra file (file339) when file339 by itself is perfectly okay.
Potential issue?
I ran
ncdump -hon files 338/339. The global attributes show the tiling structure changes right at file339. All prior files have a tiling of020x018but file339 and its successors have a tiling of030x024. Is this messing up theopen_mfdatasetstep when I try to combine all 365 files into one dataset?Beta Was this translation helpful? Give feedback.
All reactions