-
Beta Was this translation helpful? Give feedback.
Replies: 1 comment
-
|
I suppose that by "far-field" you mean grain centers and volumes, and you know the shape and size of the domain. In the standard case, you would need to define the domain size using In your case, to add the buffers, you will need to define a slightly larger domain, and to add data to the centroid/diameq file This being said, and on a more scientific front, since the grains in the buffer zones won't be "real" grains, I don't think that considering them (versus imposing boundary conditions directly to the region of interest) would add any value to the simulation. |
Beta Was this translation helpful? Give feedback.
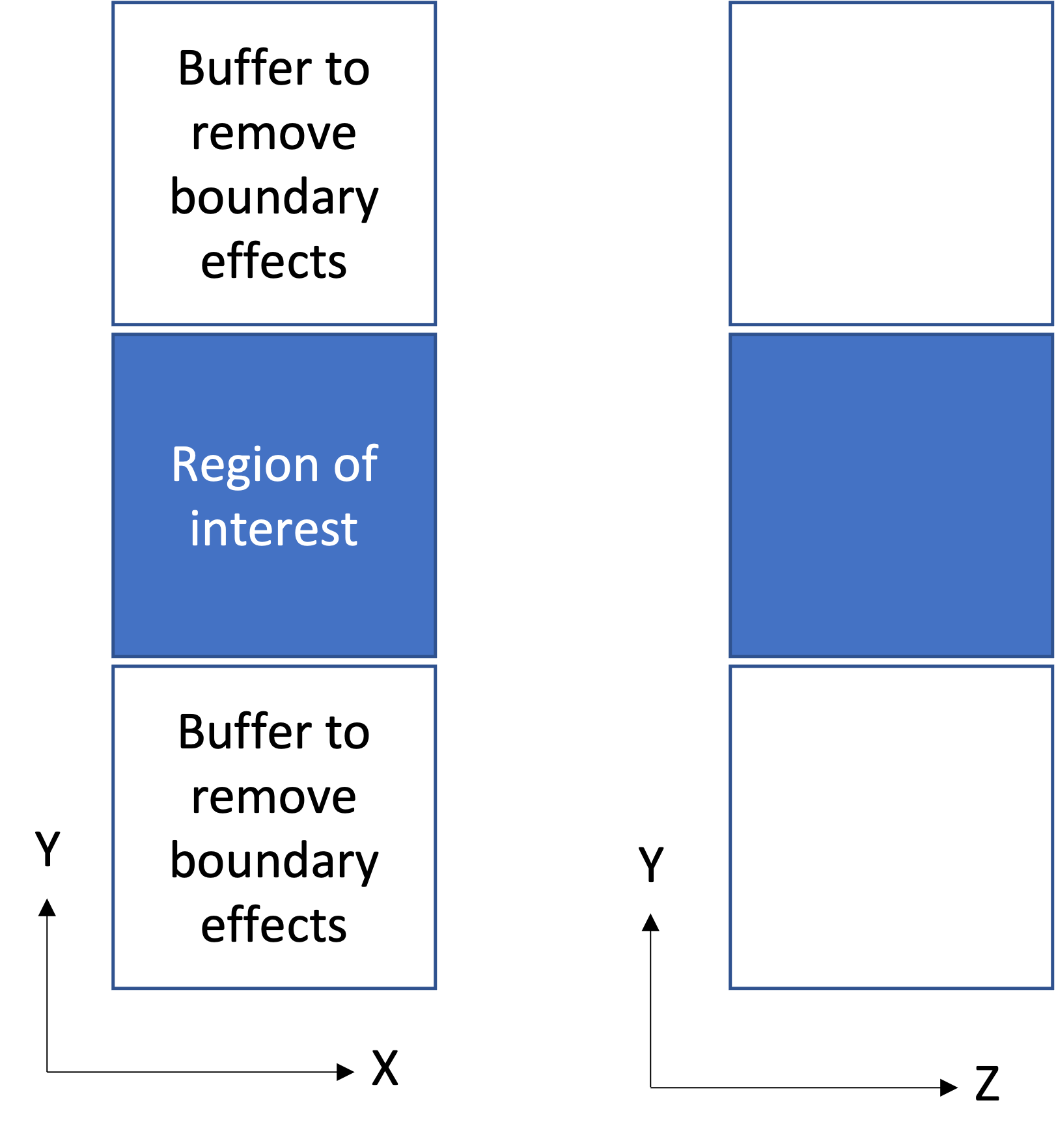
I suppose that by "far-field" you mean grain centers and volumes, and you know the shape and size of the domain.
In the standard case, you would need to define the domain size using
-domainand specify the grain centers and volumes using-morpho centroiddiameq:file(<filename>).In your case, to add the buffers, you will need to define a slightly larger domain, and to add data to the centroid/diameq file
for the grains within the buffers.
This being said, and on a more scientific front, since the grains in the buffer zones won't be "real" grains, I don't think that considering them (versus imposing boundary conditions directly to the region of interest) would add any value to the simulation.