color coded the grain #270
-
|
Hello, I would like to create a tessellation that is color-coded red for small grain, green with medium, and blue for large grain. does neper can help with that? How I can visualize the grain with these colors coding? Also how I can generate three different sizes using the same morpho. Thanks! |
Beta Was this translation helpful? Give feedback.
Replies: 2 comments 3 replies
-
|
The easiest and more versatile way is to export the cell sizes using
Not sure what you mean, but if you want the sizes to follow a unique distribution, you can typically use |
Beta Was this translation helpful? Give feedback.
-
|
Thank you, that is very helpful!
…On Thu, Jul 22, 2021 at 1:57 AM Romain Quey ***@***.***> wrote:
The easiest and more versatile way is to export the cell sizes using
-statcell, use this file to create a cell color file, and use the cell
color file in -V:
$ neper -T -n 1000 -statcell diameq
$ awk '{if ($1<0.11) print "red"; else if ($1<0.13) print "green"; else print "blue"}' n1000-id1.stcell > cell-colors
$ neper -V n1000-id1.tess -datacellcol "col:file(cell-colors)" -print img
[image: image]
<https://user-images.githubusercontent.com/2659376/126613113-9d4a416d-cbc5-41cf-894b-bd10067fff86.png>
Also how I can generate three different sizes using the same morpho
Not sure what you mean, but if you want the sizes to follow a unique
distribution, you can typically use -morpho diameq:lognormal(...), and if
you want the sizes to follow different distributions, you can sum these
distributions, as in -morpho
"diameq:lognormal(0.1,0.035)+0.2*lognormal(0.2,0.070)+0.1*lognormal(0.3,0.105)"
.
—
You are receiving this because you authored the thread.
Reply to this email directly, view it on GitHub
<#270 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AI32CHXER6HF466U5ENQQLLTY7MN7ANCNFSM5AW2TUDA>
.
|
Beta Was this translation helpful? Give feedback.

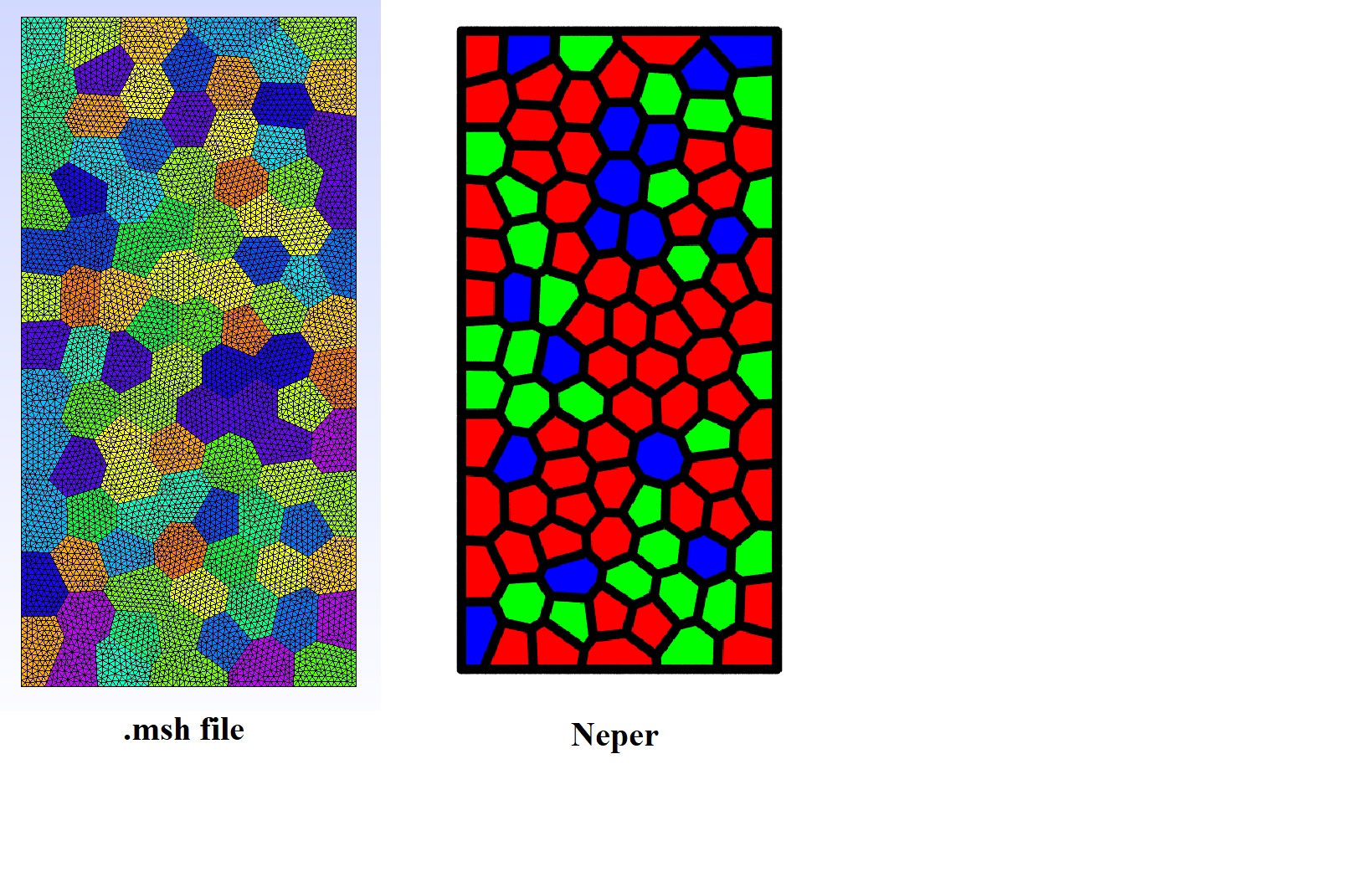
The easiest and more versatile way is to export the cell sizes using
-statcell, use this file to create a cell color file, and use the cell color file in-V:Not sure what you mean, but if you want the sizes to follow a unique distribution, you can typically use
-morpho diameq:lognormal(...), and if you want the sizes to follow different distributions, you can sum these distributions, as in-mo…