You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
Copy file name to clipboardExpand all lines: README.md
+21-26Lines changed: 21 additions & 26 deletions
Original file line number
Diff line number
Diff line change
@@ -4,9 +4,8 @@ This application creates visualizations of FASTA formatted DNA nucleotide data.
4
4
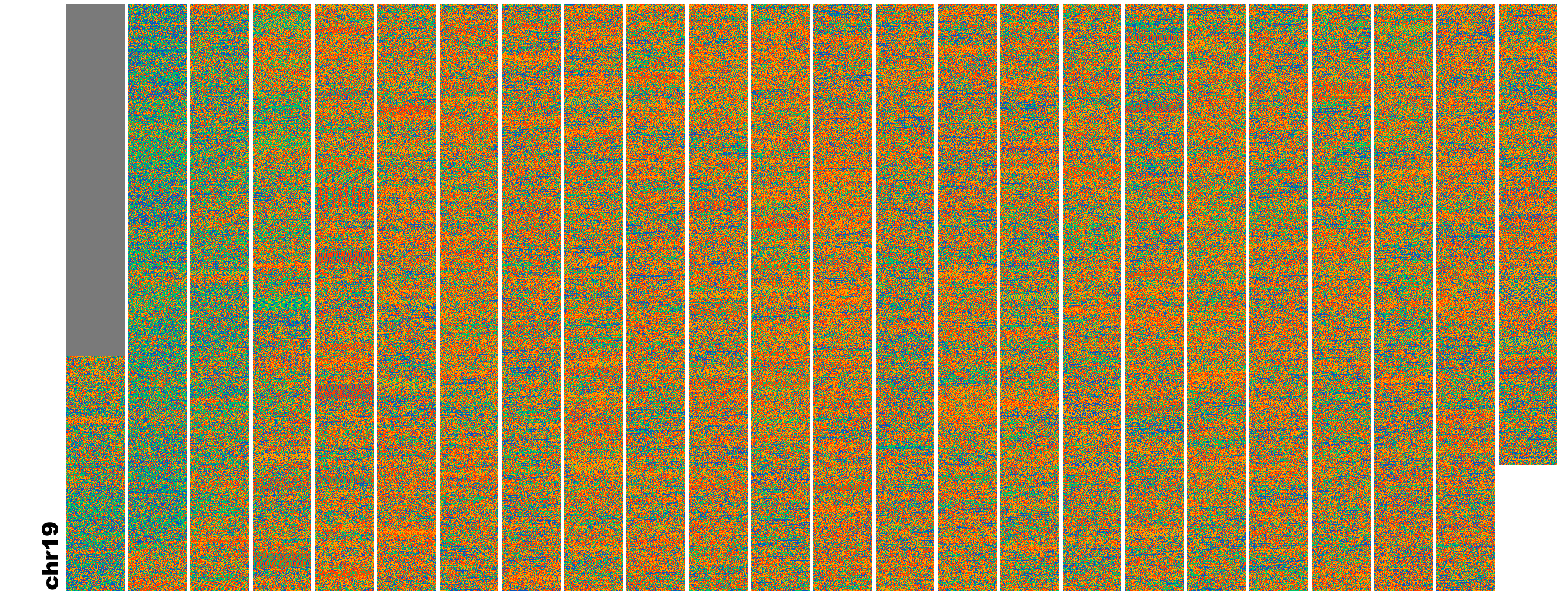
FluentDNA generates a DeepZoomImage visualizations similar to Google Maps for FASTA files.
5
5
6
6
From 2Mbp of Genomic Sequence, FluentDNA generates this image. Changes in nucleotide bias make
7
-
individual genome elements visible even without an annotation. Add your annotation files to see how
8
-
they align with the sequence features.
9
-

7
+
individual genome elements visible even without an annotation. Add your annotation files to see how they align with the sequence features.
8
+
10
9
11
10
***
12
11
## FluentDNA Quick Start
@@ -18,13 +17,12 @@ You can start using FluentDNA:
18
17
4. Your result files will be placed in the FluentDNA directory `FluentDNA/results/`. Once your private server has started,
19
18
all your results are viewable at [http://localhost:8000](http://127.0.0.1:8000).
20
19
21
-
To use FluentDNA as a python module (required for **Linux**), follow the [pip install instructions](https://github.com/josiahseaman/FluentDNA/blob/python-master/docs/installation.md).
20
+
FluentDNA can be installed as a python module (required for **Linux**).
21
+
See the [pip install instructions](https://github.com/josiahseaman/FluentDNA/blob/python-master/docs/installation.md) for more details.
22
22
```
23
-
pip install --upgrade DNASkittleUtils
24
23
pip install --upgrade FluentDNA
25
-
```
26
-
**Locating Results:** You will need to be using the same computer the server is running on. The server will not be visible
27
-
over network or internet unless your administrator opens the port.
24
+
```
25
+
**Locating Results:** You will need to be using the same computer the server is running on. The server will not be visible over network or internet unless your administrator opens the port.
28
26
29
27
***
30
28
@@ -47,12 +45,10 @@ Generating a basic visualization of a FASTA file downloaded from NCBI or another
47
45
48
46
This generates an image pyramid with the standard legend (insert image of legend) and nucleotide number display.
49
47
50
-
**Input Data Example:**
51
-
52
-
* FA File: [FluentDNA/example_data/hg38_chr19_sample.fa](https://github.com/josiahseaman/FluentDNA/blob/python-master/FluentDNA/example_data/hg38_chr19_sample.fa)
48
+
**Input Data Example:**[FluentDNA/example_data/hg38_chr19_sample.fa](https://github.com/josiahseaman/FluentDNA/blob/python-master/FluentDNA/example_data/hg38_chr19_sample.fa)
This generates a multi-scale image of the multi-part FASTA file. Note that if you don't specify `--outname=` it will default to the name of the FASTA file.
85
81
86
82
Using this simple command, FluentDNA can visualize an entire draft genome at once.
87
-
**Result:**[Ash Tree Genome (_Fraxinus excelsior_)](https://dnaskittle.com/ddvresults/dnadata/Ash%20Tree%20Genome%20-%20BATG-0_5/)
83
+
**Result:**[Ash Tree Genome (_Fraxinus excelsior_)](https://FluentDNA.com/archive/Ash%20Tree%20Genome%20-%20BATG-0_5/)
You can download the full _Gnetum montanum_ files from [Data Dryad](https://datadryad.org//resource/doi:10.5061/dryad.0vm37).
@@ -117,7 +112,7 @@ You can download the full _Gnetum montanum_ files from [Data Dryad](https://data
117
112
To visualize a multiple sequence alignment you need to use the `--layout=alignment` option to tell FluentDNA to treat each entry in a multipart fasta file as being one row of an alignment. To show many MSAs at once, just point `--fasta=` to a folder instead of a file.
118
113
119
114
**Input Data Example:**
120
-
* Folder with FA files: https://github.com/josiahseaman/FluentDNA/tree/python-master/FluentDNA/example_data/alignments
115
+
*[Folder with FA files](https://github.com/josiahseaman/FluentDNA/tree/python-master/FluentDNA/example_data/alignments)
121
116
122
117
**Important Note!** Make sure there are no other files in your folder besides sequence files. If FluentDNA decides on an unreasonably long "max width" it is because it picked up a non-sequence file in the folder.
123
118
@@ -133,10 +128,10 @@ ACTCAAAACGATCTCTCTAGGGT
133
128
134
129
This generates a multi-scale image of the multiple alignment. The multiple alignment results are sorted by gene name. For a smoother layout use `--sort_contigs` which will sort them by row count (copy number).
135
130
136
-
**Result:**[Example 7 Gene Families from Fraxinus](https://dnaskittle.com/ddvresults/dnadata/Example%207%20Gene%20Families%20from%20Fraxinus/)
131
+
**Result:**[Example 7 Gene Families from Fraxinus](https://FluentDNA.com/archive/Example%207%20Gene%20Families%20from%20Fraxinus/)
This layout allows users to check thousands of MSAs. Here we used FluentDNA to quality check the merging software for 2,961 putative gene families: [Fraxinus Homologous Gene Groups](https://dnaskittle.com/ddvresults/dnadata/Fraxinus%20Homologous%20Gene%20Groups/)
134
+
This layout allows users to check thousands of MSAs. Here we used FluentDNA to quality check the merging software for 2,961 putative gene families: [Fraxinus Homologous Gene Groups](https://FluentDNA.com/archive/Fraxinus%20Homologous%20Gene%20Groups/)
140
135
141
136
***
142
137
@@ -158,7 +153,7 @@ This generates a multi-scale image of the alignment. There are 4 columns in thi
158
153
* Column 3. Genome B (unique DNA of B)
159
154
* Column 4. Genome B (gapped entire DNA of genome B)
160
155
161
-
**Result:**[Human vs Chimpanzee_chr19 (natural colors)](https://dnaskittle.com/ddvresults/dnadata/Parallel_hg38_and_panTro5_chr19/)
156
+
**Result:**[Human vs Chimpanzee_chr19 (natural colors)](https://FluentDNA.com/archive/Parallel_hg38_and_panTro5_chr19/)
162
157

163
158
164
159
Figure 1 in the paper can be seen at Nucleotide Position 548,505 which corresponds to HG38 chr19:458,731. The difference in coordinates is due to the gaps inserted for sake of alignment.
Copy file name to clipboardExpand all lines: docs/installation.md
+5-5Lines changed: 5 additions & 5 deletions
Original file line number
Diff line number
Diff line change
@@ -1,17 +1,16 @@
1
1
This will guide you through installing FluentDNA as a python module called "FluentDNA". This is the ideal
2
2
option for developers who want to integrate or tweak FluentDNA. Non-technical users should use
3
-
a release from the [Releases Page](https://github.com/josiahseaman/FluentDNA/releases).
3
+
a release from the [Releases Page](https://github.com/josiahseaman/FluentDNA/releases).
4
4
5
5
## Quick Start
6
6
You will need:
7
-
1. Familiarity with the command line: [Windows Tutorial](https://github.com/pettarin/python-on-windows)[Mac Tutorial](http://docs.python-guide.org/en/latest/starting/install3/osx/#install3-osx)
7
+
1. Familiarity with the command line: [Windows Tutorial](https://github.com/pettarin/python-on-windows),[Mac Tutorial](http://docs.python-guide.org/en/latest/starting/install3/osx/#install3-osx)
8
8
1. Python (Windows must by Python 3.6 or newer): [Download Link](https://www.python.org/downloads/release/python-365/)
fluentdna.py will be placed in the scripts folder and accessible through PYTHONPATH, it's a good idea to add this to PATH.
25
-
The `example_data` directory will end up in your site packages, so you'll need to reference the full path.
24
+
You should be able to use `fluentdna.py` as an executable from anywhere, depending on your setup. Running `fluentdna.py` with no arguments will point you to your *example_data/* and *results/* paths in your site packages, so you'll need to reference the full path.
0 commit comments