minian for low SNR recordings made in ventral brain regions through thin relay lenses? #192
Unanswered
Gabrielle-Siemonsmeier
asked this question in
Q&A
Replies: 1 comment 3 replies
-
|
I personally don't have experience in this brain region, but I could throw in some ideas based on the info you sent:
|
Beta Was this translation helpful? Give feedback.
3 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
Hi everyone,
I'm looking to transition from MIN1PIPE to minian to process my miniscope recordings, but I'm having a hard time getting the pipeline to work for me.
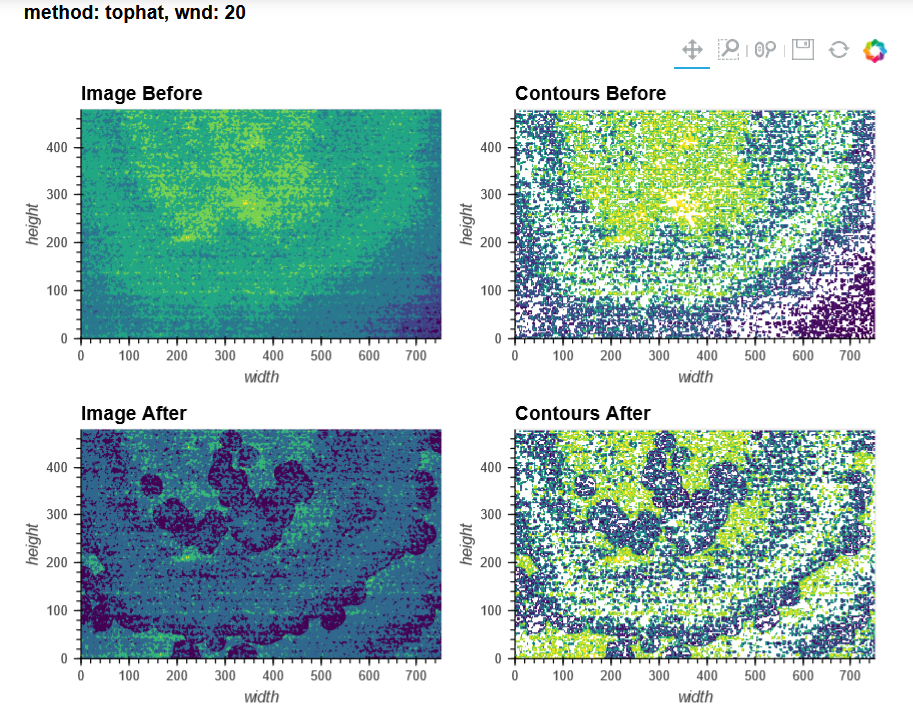
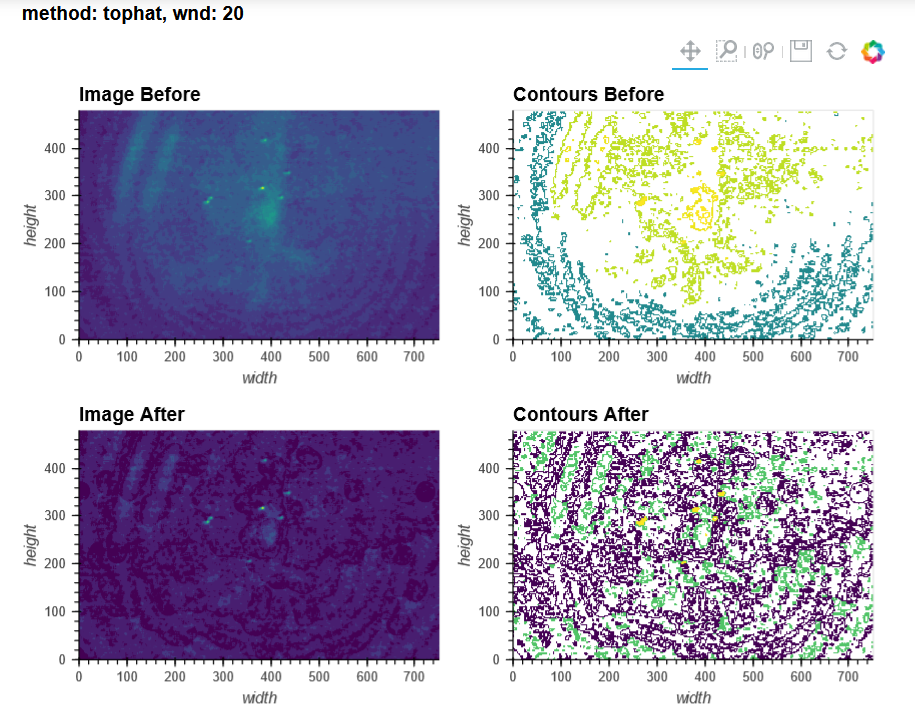
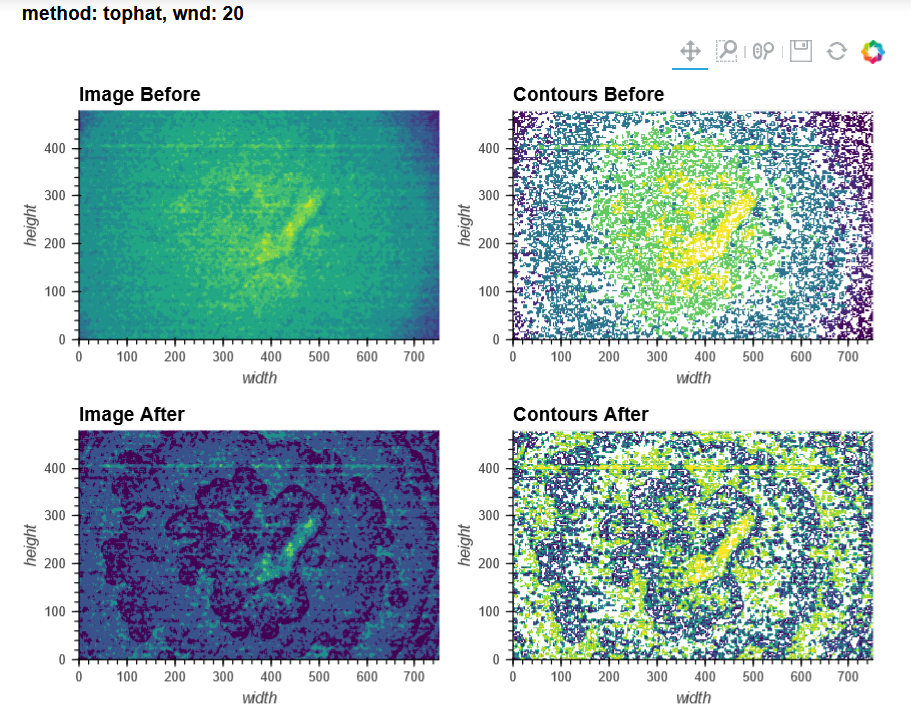
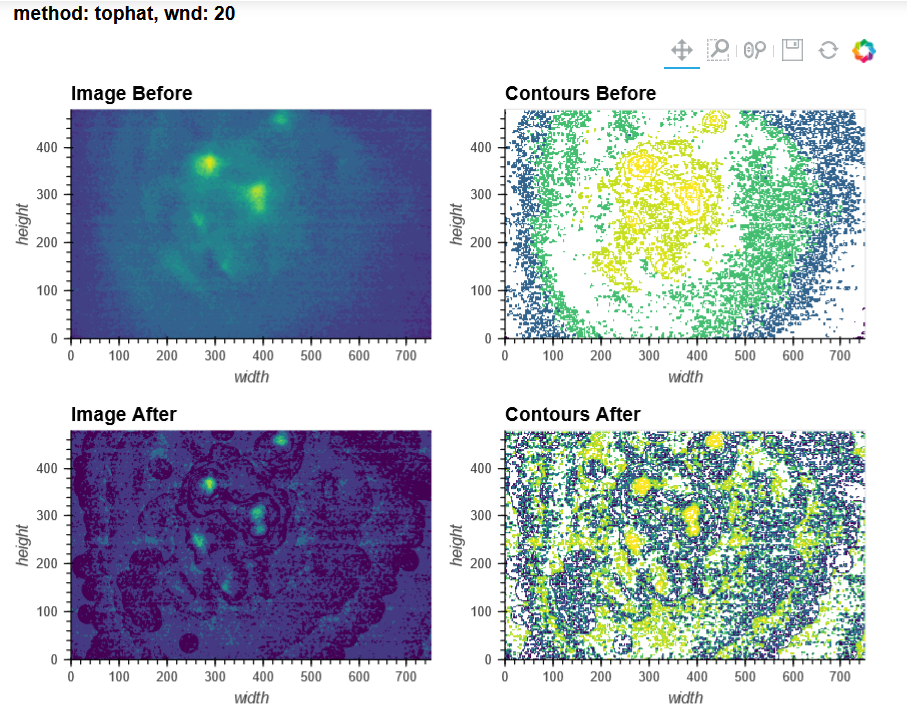
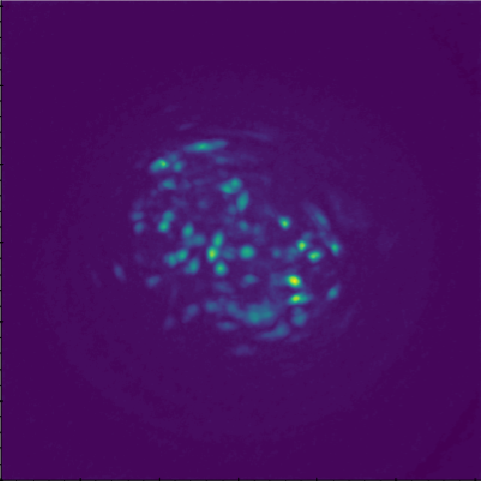
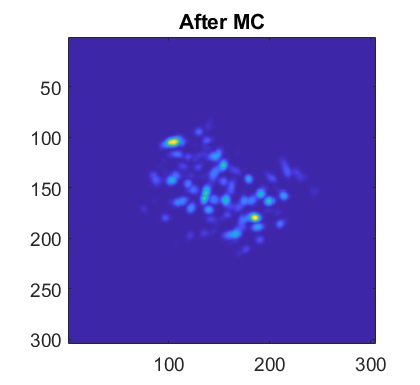
For context, I'm using V3 UCLA miniscopes to image ventral striatum (-4.6mm DV, from Bregma) in mice using 0.6 x 7.3mm relay GRIN lenses. I've imaged both CaMKIIa-GCaMP6f in WT mice and CAG-FLEX-GCaMP7s in D1-Cre mice. I've been trying to run homecage recordings (2-3min in length at 15-20fps) from several of my animals, which I had successfully processed with MIN1PIPE previously, through minian, without success so far (see attached pictures below of max projection after preprocessing and motion correction: 216, 235, 237 are WT, 289, 292, 295 are D1-Cre; sorry for how crude these are, can share other visualizations if useful).
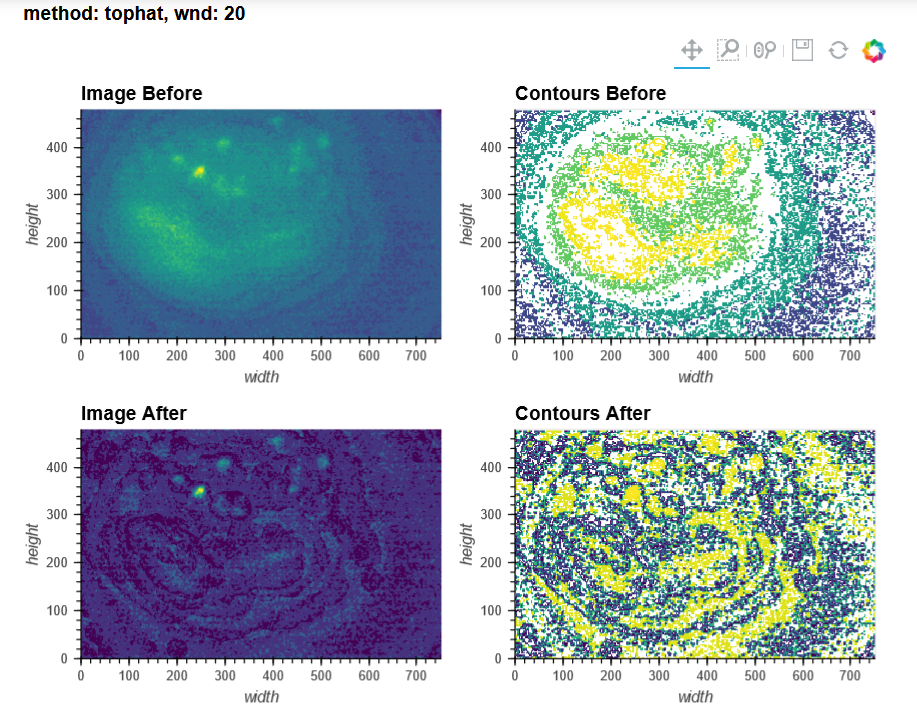
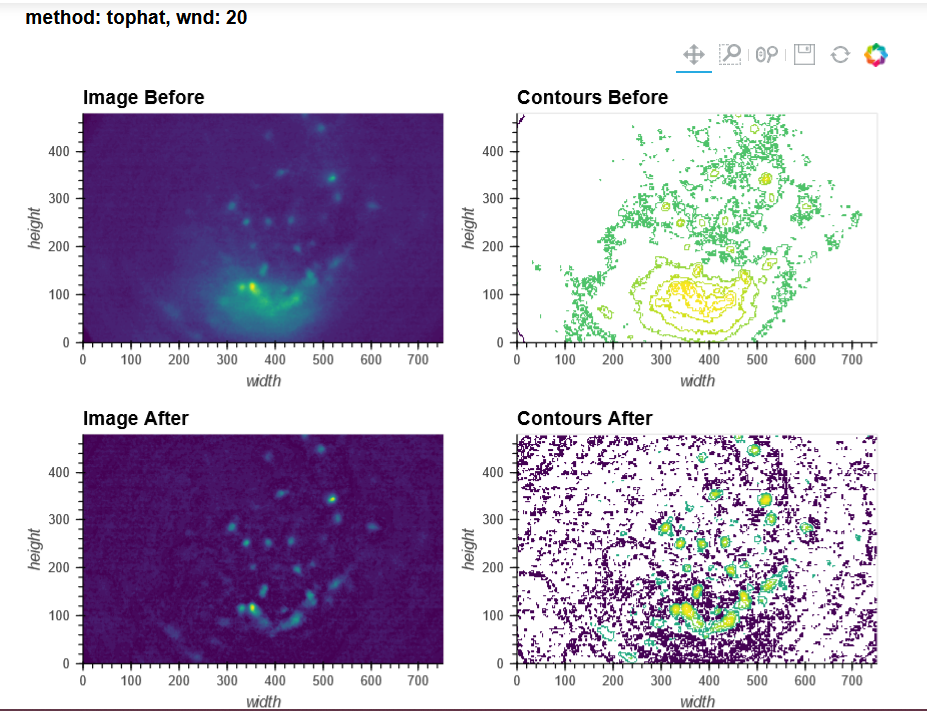
By contrast, a 50s recording made of a Cre-dependent GCaMP6f in the dorsal striatum of an A2a-Cre mouse through a 1mm diameter relay lens was processed beautifully in both pipelines (see attached picture below of max projection after preprocessing and motion correction: 3249). I reckon this recording more closely resembles the demo data, which I understood to be dorsal CA1, presumably imaged through a single 1.8mm GRIN lens (correct me if I'm wrong!)
Here's a dropbox folder containing the final minian visualization videos: https://www.dropbox.com/sh/ite4qp76xkp8ad9/AACwTt0mQi7UzDU8052DNo_Qa?dl=0 (let me know if there's any access issues!)
You can tell from the top right that motion correction is often unsatisfactory (and motion artifacts get 'baked in'), and from the bottom left and right that cell identification and trace extraction seems to fail, sometimes entirely.
The main differences between my dorsal vs. ventral striatum recordings seem to me to be the following:
In MIN1PIPE, I've used sr=0.5 (spatial downsampling by a factor of 2) and se=5 or 6 (structural element for background removal) for the dorsal striatum recording (as well as sr=1 and se=10 or 12, which yielded a number of units closer to what I obtained with minian, but a projection and neural contours that looked less accurate), and sr=0.25 (spatial downsampling by a factor of 4) and se=10 (structural element for background removal) for the ventral striatum recordings. I stuck quite closely to the default minian parameters for the dorsal striatum recording, but I haven't found a set of parameters in minian that would approximate what I obtain in MIN1PIPE for the ventral striatum recordings (but maybe I'm testing the completely wrong ranges of values?), and I have had to skip the background removal step altogether.
Bottom line, I was wondering if anyone had successfully used minian for recordings from brain regions other than dorsal CA1, and in particular, any ventral brain regions that would require thin relay lenses; or more generally, recordings with low SNR, sparse cellular activity, and/or high levels of motion. Any insight, advice, tips or tricks appreciated!
Cheers,
Gabrielle
Beta Was this translation helpful? Give feedback.
All reactions