|
7 | 7 |
|
8 | 8 | This module provides a Julia interface to the [BioMASS](https://github.com/biomass-dev/biomass) parameter estimation. |
9 | 9 |
|
10 | | - |
11 | | - |
12 | | -## Features |
| 10 | +## Installation |
13 | 11 |
|
14 | | -BioMASS.jl supports: |
| 12 | +The package is a registered package, and can be installed with `Pkg.add`. |
15 | 13 |
|
16 | | -- parameter estimation of ODE/DDE models |
17 | | -- visualization of simulation results |
18 | | -- bifurcation analysis |
| 14 | +```julia |
| 15 | +julia> using Pkg; Pkg.add("BioMASS") |
| 16 | +``` |
19 | 17 |
|
20 | | -## Usage |
| 18 | +or through the `pkg` REPL mode by typing |
21 | 19 |
|
22 | | -### Parameter estimation |
| 20 | +``` |
| 21 | +] add BioMASS |
| 22 | +``` |
23 | 23 |
|
24 | | -```julia |
25 | | -using BioMASS |
| 24 | +## Example |
26 | 25 |
|
27 | | -model = Model("./examples/fos_model"); |
| 26 | +### Model development |
28 | 27 |
|
29 | | -# Estimate unknown model parameters against experimental observations. |
30 | | -optimize(model, 1, max_generation=20000, allowable_error=0.5) |
| 28 | +This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model. [`pasmopy.Text2Model`](https://pasmopy.readthedocs.io/en/latest/model_development.html) allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model. |
31 | 29 |
|
32 | | -# Save simulation results to figure/ in the model folder |
33 | | -run_simulation(model, viz_type="best", show_all=true) |
| 30 | +Prepare a text file describing the biochemical reactions (e.g., `michaelis_menten.txt`) |
34 | 31 | ``` |
| 32 | +E binds S <--> ES | kf=0.003, kr=0.001 | E=100, S=50 |
| 33 | +ES dissociates to E and P | kf=0.002, kr=0 |
35 | 34 |
|
36 | | -### Conversion of optimized parameters into BioMASS format |
| 35 | +@obs Substrate: u[S] |
| 36 | +@obs E_free: u[E] |
| 37 | +@obs E_total: u[E] + u[ES] |
| 38 | +@obs Product: u[P] |
| 39 | +@obs Complex: u[ES] |
37 | 40 |
|
38 | | -```julia |
39 | | -param2biomass("./examples/fos_model") |
| 41 | +@sim tspan: [0, 100] |
40 | 42 | ``` |
41 | 43 |
|
42 | | -## Installation |
43 | | - |
44 | | -The package is a registered package, and can be installed with `Pkg.add`. |
| 44 | +Convert the text into an executable model |
45 | 45 |
|
46 | | -```julia |
47 | | -julia> using Pkg; Pkg.add("BioMASS") |
| 46 | +```shell |
| 47 | +$ python # pasmopy requires Python 3.7+ |
| 48 | +``` |
| 49 | +```python |
| 50 | +>>> from pasmopy import Text2Model |
| 51 | +>>> description = Text2Model("michaelis_menten.txt", lang="julia") |
| 52 | +>>> description.convert() # generate 'michaelis_menten_jl/' |
48 | 53 | ``` |
49 | 54 |
|
50 | | -or through the `pkg` REPL mode by typing |
| 55 | +Simulate the model using BioMASS.jl |
51 | 56 |
|
| 57 | +```shell |
| 58 | +$ julia |
52 | 59 | ``` |
53 | | -] add BioMASS |
| 60 | +```julia |
| 61 | +using BioMASS |
| 62 | + |
| 63 | +model = Model("./michaelis_menten_jl"); |
| 64 | +run_simulation(model) |
54 | 65 | ``` |
55 | 66 |
|
56 | | -## References |
| 67 | +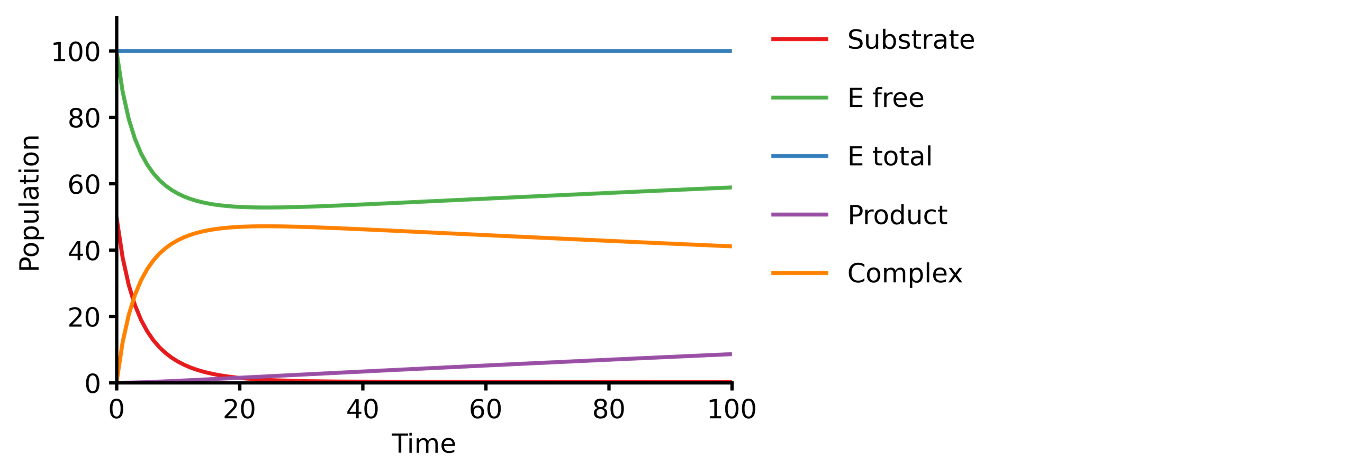 |
| 68 | +### Parameter estimation |
57 | 69 |
|
58 | | -- Nakakuki, T. _et al._ Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. _Cell_ **141**, 884–896 (2010). https://doi.org/10.1016/j.cell.2010.03.054 |
| 70 | +```julia |
| 71 | +using BioMASS |
59 | 72 |
|
60 | | -- Inoue, K. _et al._ Oscillation dynamics underlie functional switching of NF-κB for B-cell activation. _npj Syst. Biol. Appl._ **2**, 16024 (2016). https://doi.org/10.1038/npjsba.2016.24 |
| 73 | +model = Model("./examples/fos_model"); |
61 | 74 |
|
62 | | -- Yao, G., Lee, T. J., Mori, S., Nevins, J. R. & You, L. A bistable Rb-E2F switch underlies the restriction point. _Nat. Cell Biol._ **10**, 476–482 (2008). https://doi.org/10.1038/ncb1711 |
| 75 | +# Estimate unknown model parameters against experimental observations. |
| 76 | +optimize(model, 1, max_generation=20000, allowable_error=0.5) |
| 77 | + |
| 78 | +# Save simulation results to figure/ in the model folder |
| 79 | +run_simulation(model, viz_type="best", show_all=true) |
| 80 | + |
| 81 | +# Convert optimization results into BioMASS format |
| 82 | +param2biomass("./examples/fos_model") |
| 83 | +``` |
| 84 | + |
| 85 | +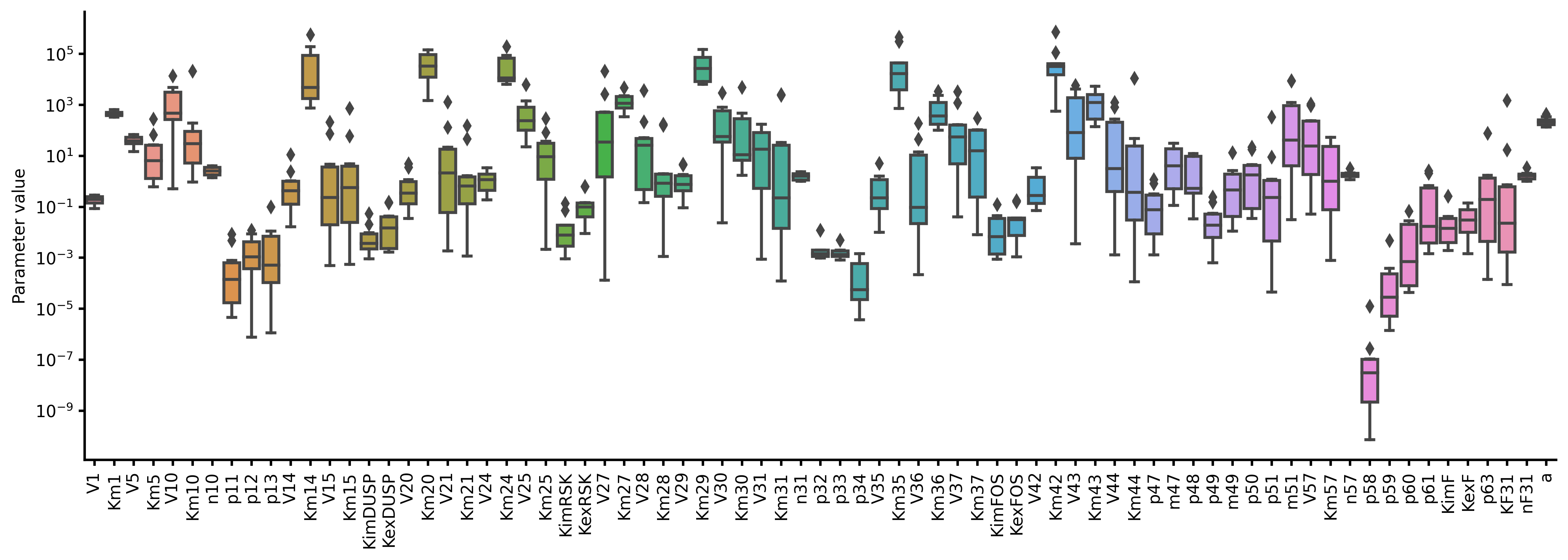 |
| 86 | + |
| 87 | +## References |
63 | 88 |
|
64 | | -- Barr, A. R., Heldt, F. S., Zhang, T., Bakal, C. & Novák, B. A Dynamical Framework for the All-or-None G1/S Transition. _Cell Syst._ **2**, 27–37 (2016). https://doi.org/10.1016/j.cels.2016.01.001 |
| 89 | +- Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. _Cancers_ **12**, 2878 (2020). https://doi.org/10.3390/cancers12102878 |
65 | 90 |
|
66 | | -- Rata, S. _et al._ Two Interlinked Bistable Switches Govern Mitotic Control in Mammalian Cells. _Curr. Biol._ **28**, 3824-3832.e6 (2018). https://doi.org/10.1016/j.cub.2018.09.059 |
| 91 | +- Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. _iScience_ (2022). https://doi.org/10.1016/j.isci.2022.103944 |
67 | 92 |
|
68 | 93 | ## License |
69 | 94 |
|
|
0 commit comments