Replies: 3 comments 30 replies
-
|
Can you explain what you mean by "% error expression"? |
Beta Was this translation helpful? Give feedback.
9 replies
-
|
I think I see the problem You should be using left_join (or something similar) where you retain only annotation information that matches data table with expression |
Beta Was this translation helpful? Give feedback.
7 replies
-
|
Put a link to the GitHub repo where you are working.. that would be easiest way to understand and troubleshoot |
Beta Was this translation helpful? Give feedback.
14 replies
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
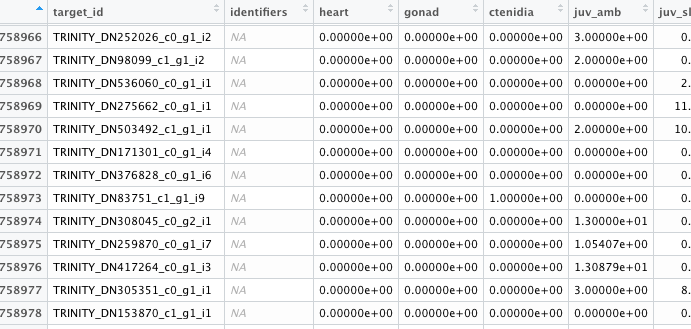
Lets say my data looks like this table after the count data has been aligned with the blast data. In order to calculate % error expression, my first reaction would be to calculate % NA in the Identifier column, but sometimes there is count data without an identifier.
My second guess would be to filter the tpm so only values > 0 are counted, then calculate the % that has any value at all, but would some values be NA and some 0?
I don't think it makes any sense to calculate % error expressed using the Target_ID column since most of them have values but sometimes there are identifiers that don't have a corresponding Target_ID.
If I use the R function complete.cases, it removed any row with NAs which would delete all rows 1-3 in this example, and in my mind if there is count data but no identifiers, wouldn't that mean the gene exists but we haven't identified it yet? Any help with this data wrangling would be appreciated.
Beta Was this translation helpful? Give feedback.
All reactions