-
|
I'm mired in R list junk and am in desperate need of someone to walk through how to deal with/understand lists. I find it confusing on how to deal with lists containing different type of objects (e.g. a list of data frames) and how to work with them. The end goal is extract/combine data and determine row-wise coefficients of variation. Are there any list experts who'd be willing/interested in spending some time walking through my project and providing me with some guidance/explanation/help? Science hour would be ideal. Otherwise, maybe we can explore some of this at the next lab meeting. Or, "worst case" I'm also up for a dedicated session specifically for this at some other time. In the mean time, I'll continue to do some reading/practicing on the topic and see if I can get a better grasp of things. The other benefit of the session might also reveal some different approaches that other people would take which would be significantly less confusing!!! For kicks, I'll post some of the stuff I'm working on in this discussion, in case anyone wants to peruse it. However, it quickly gets a bit complicated to explain without walking through the entire process, thus my interest in discussing via video chat. |
Beta Was this translation helpful? Give feedback.
Replies: 5 comments 3 replies
-
|
Git Repo: https://github.com/epigeneticstoocean/2018_L18-adult-methylation R Markdown file: https://github.com/epigeneticstoocean/2018_L18-adult-methylation/blob/main/code/ballgown_analysis.Rmd I'm working with the gene expression analysis package Part of code that I'm trying to wrangle begins here:
This is part where it is difficult to get the information into a text format without perusing the data with someone else present. Since it's an object, there's a lot of information associated with it that I can't easily copy/paste here. Next, I can easily extract transcript expression (FPKM) data from that subset of samples (for looping purposes, I pop this info into a list): Here's what one entry in the list looks like: treatment_fpkm_list[[1]] FPKM.S13M FPKM.S64M FPKM.S6M FPKM.S7M
1 63.109000 1161.543557 110.668782 239.215973
2 158.217649 82874.949689 487.061698 2602.800279
3 72.036519 963.764924 93.048271 241.462390
4 55.182350 107.488847 52.721938 127.347810
5 97.711458 154.764867 84.386166 145.411473I also have a data frame where the I'd like to "merge" this data frame and the info from the list above, keeping only the columns in that list and adding Or, a different approach would be to use the contents of the list to select the desired columns from the data frame. Actually, this might be a better approach? Any suggestions for this step? |
Beta Was this translation helpful? Give feedback.
-
|
I'm getting closer. Need/want to add column to items in list from a vector of gene names. Here's what I did: # Add column called "gene_name", comprised of gene names, to existing list item
> treatment_fpkm_list[[1]] <- cbind(treatment_fpkm_list[[1]], gene_name = whole_tx_table_gene.names)
> treatment_fpkm_list[[1]]This adds a column and makes the list item look like this: FPKM.S13M FPKM.S64M FPKM.S6M FPKM.S7M gene_name
1 "63.109" "1161.543557" "110.668782" "239.215973" "COX1"
2 "158.217649" "82874.949689" "487.061698" "2602.800279" "rna-NC_007175.2:1710..8997"
3 "72.036519" "963.764924" "93.048271" "241.46239" "COX3"
4 "55.18235" "107.488847" "52.721938" "127.34781" "rna-NC_007175.2:3430..3495"
5 "97.711458" "154.764867" "84.386166" "145.411473" "rna-NC_007175.2:3499..3567"
6 "82.522413" "154.972338" "67.927814" "120.346216" "rna-NC_007175.2:3578..3646" |
Beta Was this translation helpful? Give feedback.
-
|
Have you tried the ‘bind_rows’ function? For example I used it in the below code to combine data frames saved to the list object “DML.features”, except for the 7th df in the list. DML.features.df <- bind_rows(DML.features[-7]) |
Beta Was this translation helpful? Give feedback.
-
|
I have figured out some stuff in |
Beta Was this translation helpful? Give feedback.
-
|
Thanks to those that chimed in. Even though I made this a Discussion about list manipulation, it was really a specific issue in manipulating For posterity, I'll post the true underlying issue that was screwing me up. Let's say one has created a
If one wants to extract gene info, one would run this command:
Although these commands, have virtually identical syntax, there is a critical difference in what is output.
I was trying to manipulate the outputs from both commands, expecting a Once I figured out that |
Beta Was this translation helpful? Give feedback.
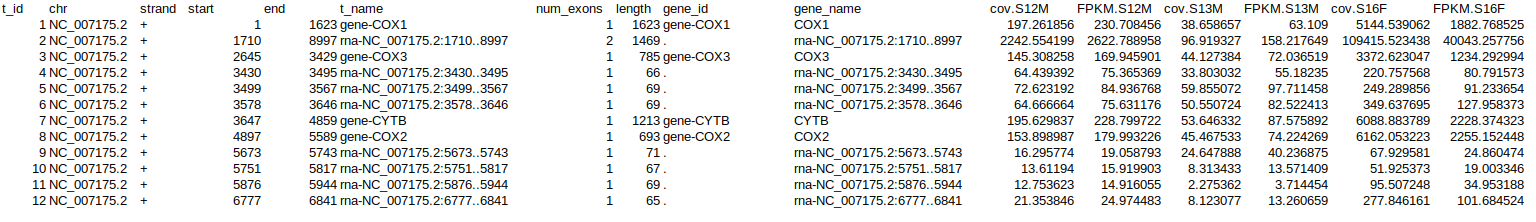
Thanks to those that chimed in. Even though I made this a Discussion about list manipulation, it was really a specific issue in manipulating
ballgownobjects and tying into using therowcvs()function.For posterity, I'll post the true underlying issue that was screwing me up.
Let's say one has created a
ballgownobject, calledbg. To extract transcript info, one would run this command:texpr(bg)If one wants to extract gene info, one would run this command:
gexpr(bg)Although these commands, have virtually identical syntax, there is a critical difference in what is output.
texpr(bg)spits out aballgownobject.gexpr(bg)spits out a matrix.I was trying to manipulate the outputs from bot…