-
|
Starting this discussion for questions about @kubu4's workflow: https://nbviewer.jupyter.org/github/RobertsLab/code/blob/master/notebooks/sam/20210601_ssal_gff-annotations.ipynb I'm running the Uniprot mapping in this Jupyter notebook with this perl script. I got the following error: What is
|
Beta Was this translation helpful? Give feedback.
Replies: 3 comments 12 replies
-
|
You may need to use |
Beta Was this translation helpful? Give feedback.
-
|
After perl module installation and running the code, I get a sever closed connection error: How do I access the server? |
Beta Was this translation helpful? Give feedback.
-
|
Looks like using the protein IDs from the protein annotation FastA in Uniprot gives results! I'm going to try working from that file and isolate the IDs to see if that works. If there's a way to modify the perl script that's great, but if not I'll just use the GUI for now. |
Beta Was this translation helpful? Give feedback.
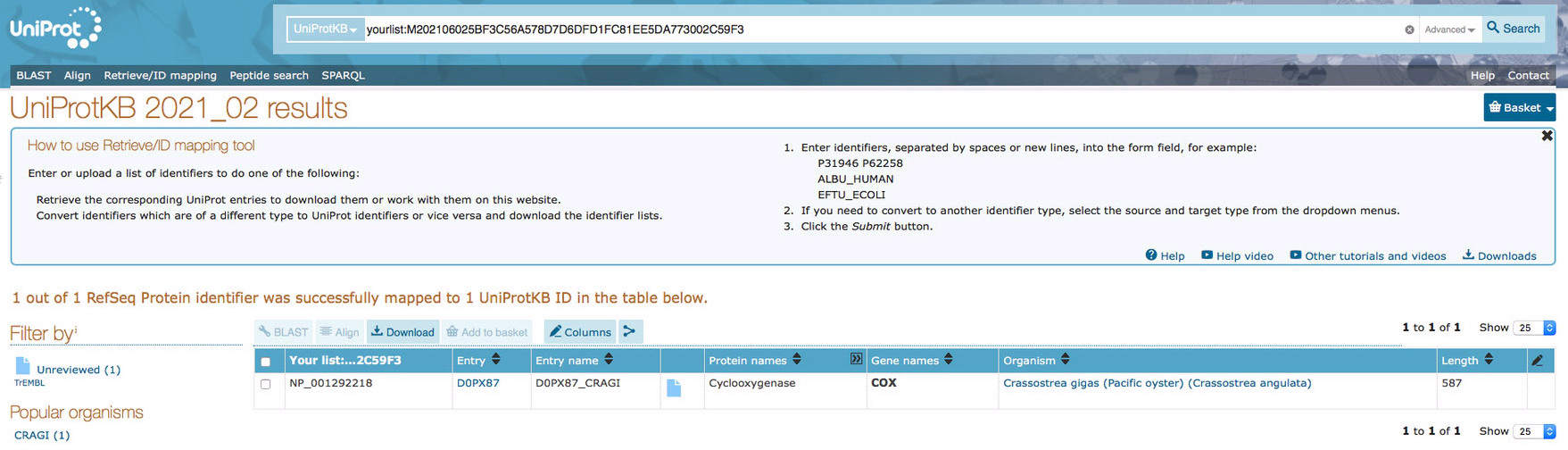
Looks like using the protein IDs from the protein annotation FastA in Uniprot gives results! I'm going to try working from that file and isolate the IDs to see if that works. If there's a way to modify the perl script that's great, but if not I'll just use the GUI for now.