-
|
I want to count the number of CG motifs in the Roslin genome. I used the following command to do so: ~4 million CpGs seems low (C. virginica has ~14 million). When I look at edited to include |
Beta Was this translation helpful? Give feedback.
Replies: 5 comments 17 replies
-
|
This will miss instances when a CG occurs at a newline. e.g.: |
Beta Was this translation helpful? Give feedback.
-
|
I think the approach is to convert the FastA into a tab-delimited file. I use seqkit to do this, usually. Then, count with awk (possibly need to use gawk, to accommodate large number of fields). E.g. # Use seqkit to convert FastA to tab-delimited and print sequence length
${seqkit} fx2tab \
--length \
${fasta} \
> ${fasta}.tab
# Print only sequences to new file
gawk '{ print $2 }' ${fn}_tab > ${fasta}.tab2
# Delimit sequences on CGs and print the number of fields minus 1 to get the number of CGs present.
gawk -F\[Cc][Gg] '{print NF-1}' ${fasta}.tab2 > CG |
Beta Was this translation helpful? Give feedback.
-
|
@sr320 How do you usually generate the CG motif files for genomes? Perhaps that's what I should be doing, so I have that as an IGV track |
Beta Was this translation helpful? Give feedback.
-
|
Also note I will separately be dealing with the general ability to count CG at #1207 |
Beta Was this translation helpful? Give feedback.
-
|
Here's my CG motif track (will add to the handbook): http://owl.fish.washington.edu/halfshell/genomic-databank/cgigas_uk_roslin_v1_fuzznuc_CGmotif.gff Similar count to the 13080574 from Looks the same as Steven's: |
Beta Was this translation helpful? Give feedback.





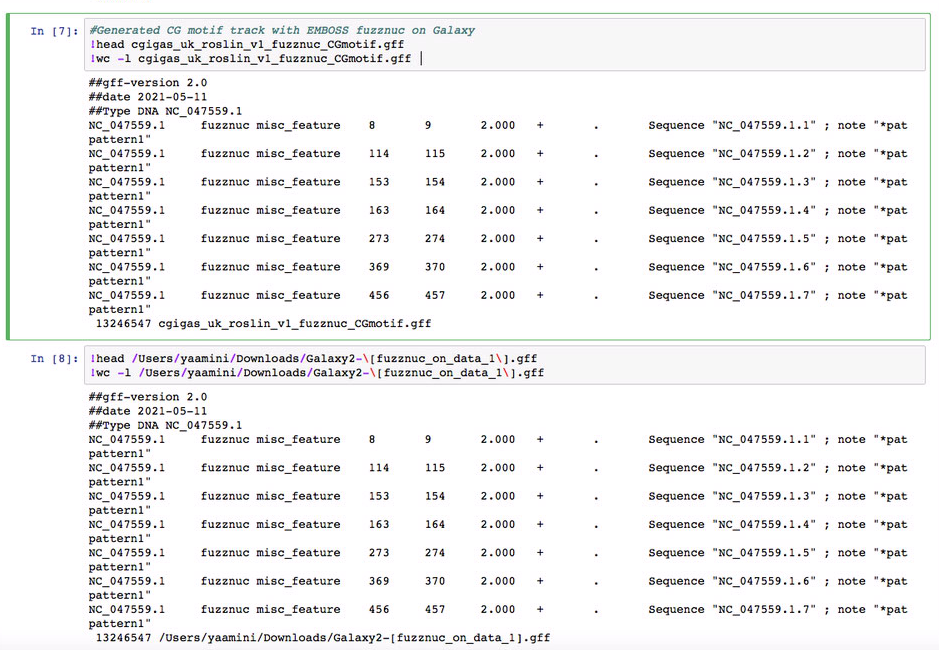
Here's my CG motif track (will add to the handbook): http://owl.fish.washington.edu/halfshell/genomic-databank/cgigas_uk_roslin_v1_fuzznuc_CGmotif.gff Similar count to the 13080574 from
fgrepLooks the same as Steven's: