-
Notifications
You must be signed in to change notification settings - Fork 48
Streaming Processing
When we develop these functionalities we are thinking about facilities. Currently, scipion is being used every day in several facilities in Europe, and more are joining from US and Canada. If you are running a Cryo EM facility and want more info, please Contact us. We will be happy to help you run Scipion there.
In the last months, we have been working on an extension of Scipion to process data in streaming, i.e, at the same time movies (or micrographs) are coming from the microscope PC. This allows to overlap computing time with the acquisition (reducing computational needs) and also to detect problems at early stages. This idea is implemented in different labs mainly by using custom-made scripts. The advantage of our Scipion solution is that you have the usual flexibility to choose what operations to do and the traceability to re-do some of the steps later. It is basically the same Scipion interface with one key change: the output is produced as soon as the first element is available, and it is later updated with new output elements. This allows concatenating several operations before the first one is completed.
On top of that, we have added the concept of monitors, the special protocols that constantly check how the execution of other protocols is going. We have developed several GUIs that are refreshed periodically and produce a graphical summary (e.g, CTF defocus values, system load etc). A summary is also generated in HTML format that can be easily copied to a public website to provide access for external users. (See an example here).
NOTE: This information is only available since v1.2-Caligula version.
To run the demo as it is you need to have installed:
-
motioncor2
-
ctffind4
-
eman
-
jmbFalconMovies dataset (for v1.2-Caligula version) or
-
relion13_tutorial dataset (for later versions and devel branch)
scipion install motioncor2 ctffind4 eman scipion testdata --download jmbFalconMovies relion13_tutorial
Notice that motioncor2 needs GPU acceleration.
After installing all the requirements you just have to run:
scipion demo
This will pop up a small wizard like the one below ready to go.
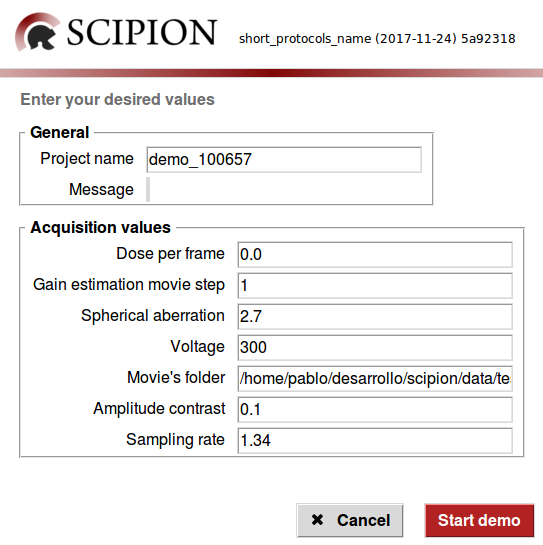
You can leave all the displayed data untouched since it goes right with the test data. Once you click on the "Start demo" button scipion should appear with the new project loaded running in streaming mode:

Import is already importing files and the rest are scheduled. As soon as there is any input available, the protocols will start processing it and making it available for the next protocol in line.
The "Monitor summary" is monitoring the progress and generating an HTML report with the outcome of the data.
To see the HTML summary report from Scipion you must first select (click) on the Summary monitor box, and once it is selected, click on the "Analyze result" button (down-right). A window like this will pop up:

Then click on the "Open HTML report" button. A browser will show you something like (this)
There are a couple of useful scripts that might be useful to start scipion right after the acquisition has started. In summary, you may want to somehow follow this steps:
-
Ask for some basic input: In our experience, so far, there is still the need of asking the user/microscopist for some basic data before Kicking off scipion. This is the case for sampling rate, or dose or even based on the flexibiliy you want to offer in your pipeline many other options like "use Gctf" or "use CTFfind4" or both….this is really up to you and Scipion does not implement any of this. The mechanism to ask for is not part of scipion, although there are some sample scripts that could be useful as an staring point, but we doubt that will be perfect as they are for your case. You will need to adapt them to your needs. The demo script (scripts/scipionbox_wizard_demo.py) might be the simplest one and does not require any config file, but it is very tied to demo purposes. Other scripts are similar to real scripts used in facilities, like (scripts/cipionbox_wizard_scilifelab.py) and are based on TK GUI. In some other facilities, the have a webserver eith an HTML form to request for this data. It is really up to your pipeline design.
-
Create the project using the input: the second step would be to use those values captured and create a scipion project accordingly. In this case you have 2 options:
-
A.- Use SCIPION as an API to create the project following your pipeline design. This is the case for scripts/cipionbox_wizard_scilifelab.py.
-
B.- Generate a "json" file, usually based on a template, that will be completed with the data requested in the previous step. This is the case of the demo script(scripts/scipionbox_wizard_demo.py). Is in this case that, once you have your "workflow.json" you can run a script to create the project based on that workflow like:
scipion python scripts/create_project.py name="session1234" workflow="path/to/your/workflow.json"
-
-
The final step would be to "Start" all protocols to kick off the processing: For the B case, there is another script that will do that:
scipion python scripts/schedule_project.py name="session1234"+
If using scipion as an API, you may want to Start the protocols ate the same step (A case)
After this, you will have Scipion up and running actively looking for new acquisitions and following the steps in your customized workflow.